| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,305,850 – 17,305,944 |
| Length | 94 |
| Max. P | 0.566626 |

| Location | 17,305,850 – 17,305,944 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.50326 |
| G+C content | 0.46814 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17305850 94 + 22422827 ------UGAAGAUCGGUGCCGUUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------(((.((((.(((((((........)))))))...(((((....(((((.((......)).))-))).....))))))))))))(-(......)).. ( -31.80, z-score = -2.25, R) >droPer1.super_18 476674 94 + 1952607 ------UGGGGUUGGGUGCUGCUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------((..(((..(((((.((((......((((((...))))))...(((((.((......)).))-))))))).))))))))..))(-(......)).. ( -32.20, z-score = -1.30, R) >dp4.chrXL_group1e 1635811 94 + 12523060 ------UGGGGUUGGGUGCUGCUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------((..(((..(((((.((((......((((((...))))))...(((((.((......)).))-))))))).))))))))..))(-(......)).. ( -32.20, z-score = -1.30, R) >droAna3.scaffold_13248 1705876 99 - 4840945 GAGAGACACGUCCUGGUGCUGUUUGGAUUUAUGGCUCACUGGGC-AACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGUGAUUUCGG-CUAAAUUGCAA ..((((((((((((.(.((((.(((..((((((((.....((..-..))))))...))))..)))..)-)))).)).)))))).)))).(-(......)).. ( -28.20, z-score = -0.76, R) >droEre2.scaffold_4690 7647001 94 + 18748788 ------UGAAGAUCGGUGCUGUUCGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------(((.((((.(((((((........)))))))...(((((....(((((.((......)).))-))).....))))))))))))(-(......)).. ( -28.90, z-score = -1.29, R) >droYak2.chrX 15925393 94 + 21770863 ------UGAAGAUCGGUGCUGUUCGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------(((.((((.(((((((........)))))))...(((((....(((((.((......)).))-))).....))))))))))))(-(......)).. ( -28.90, z-score = -1.29, R) >droSec1.super_17 1152609 94 + 1527944 ------UGAAGAUCGGUGCCGUUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------(((.((((.(((((((........)))))))...(((((....(((((.((......)).))-))).....))))))))))))(-(......)).. ( -31.80, z-score = -2.25, R) >droSim1.chrX 3433175 94 - 17042790 ------UGAAGAUCGGUGCCGUUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA ------(((.((((.(((((((........)))))))...(((((....(((((.((......)).))-))).....))))))))))))(-(......)).. ( -31.80, z-score = -2.25, R) >droGri2.scaffold_14853 1003421 93 - 10151454 -------UGGAGUUGGUGCCACUUGGAUUUAUGAUGCGCUGCGCCAACUGGCAAUUUAAAGUCAAUUU-GGCCAAGAGGCGCGACUUCAA-CUAAAUUGCAA -------((((((((.((((.(((((.....((((((...))(((....)))........))))....-..))))).)))))))))))).-........... ( -30.40, z-score = -1.80, R) >droMoj3.scaffold_6473 1504223 86 - 16943266 --------------GGUGCCGCUUGGAUUUAUGAUGCGCUGCGCCAACUGCCAAUUUAAAGGCAAUUU-AGCCAAGAGGCGCGAUUGCAG-CUAAAUUGCAA --------------((((((((.............)))..)))))...(((.((((((...((((((.-.(((....)))..))))))..-.))))))))). ( -27.62, z-score = -0.42, R) >droVir3.scaffold_12970 6167953 86 + 11907090 --------------GGUGCCGUUUGGAUUUAUGAUGCGCUGUGCCAACUGCCAAUUUAAAGGCAAUUU-GGCCAAGAGGCGCGAUUUCAG-CUAAAUUGCAA --------------..(((.((((((.....(((((((((..(((((.((((........))))..))-))).....))))))...))).-)))))).))). ( -29.10, z-score = -1.49, R) >droWil1.scaffold_180702 468434 78 + 4511350 -----------------------UGGAUUUAUGGUGAGCAGCGCCAACUGCCAAUUUAAAGACAAUUU-GGCCAAGAGGCGAGAUUUCAGACUAAAUUGCAA -----------------------........(((((.....)))))..(((.((((((..((.(((((-.(((....))).)))))))....))))))))). ( -18.20, z-score = -0.89, R) >apiMel3.Group3 10153656 95 + 12341916 ------UUAAAAACCGUGGAAUCGCGAUGUUUAUCGUUUAAUCGAAUCCUUUAAUCGCGCACCAGUUUCAGAAAGGAUACGUGGCGCGCG-UGCAGGUGUAC ------......(((..(((.(((((((....))))......))).)))......(((((.(((((.((......)).)).))).)))))-....))).... ( -24.90, z-score = -0.35, R) >consensus ______UGAAGAUCGGUGCCGUUUGGAUUUAUGGCGCACAGCGCCAACUGCCAAUUUAAAGACAAUUU_GGCCAAGAGGCGCGAUUUCAG_CUAAAUUGCAA ........................((.....((((((...))))))....))..................(((....)))(((((((......))))))).. (-14.05 = -14.07 + 0.02)


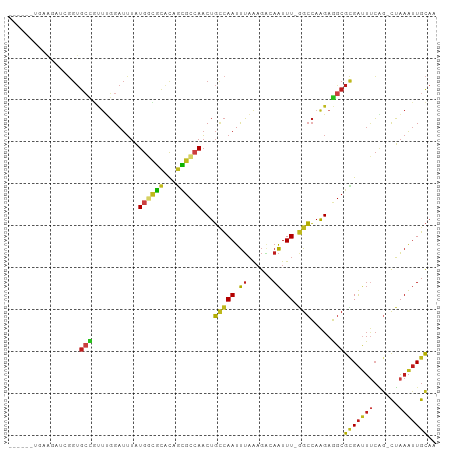
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:33 2011