| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,305,018 – 17,305,126 |
| Length | 108 |
| Max. P | 0.536600 |

| Location | 17,305,018 – 17,305,126 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Shannon entropy | 0.37885 |
| G+C content | 0.50050 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
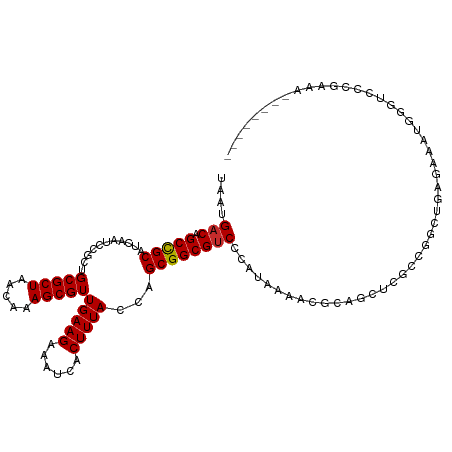
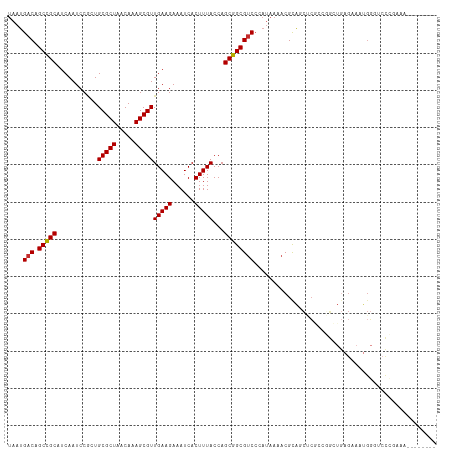
>dm3.chrX 17305018 108 + 22422827 UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCAAUAAAACGCAGCUCGCCGGCUGAGAAAUGGAUCCCGAAAA------- ......(((((((.......(((((((((.....)))).(((((......))))).)))))((((........)))).....).))))))..................------- ( -28.70, z-score = -1.10, R) >droAna3.scaffold_13248 1704381 87 - 4840945 UAAUGACAGCCGCAUCAAUCCGGUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGCAGUCCCGAAA---------------------------- ....(((.(((((...........(((((.....)))))(((((......)))))...)))))))).....................---------------------------- ( -20.10, z-score = -1.12, R) >droEre2.scaffold_4690 7645990 108 + 18748788 UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGCAGCUCGCCGGCUGAGAAAUGGGUCCCGAAAA------- ........(((((...........(((((.....)))))(((((......)))))...)))))(.(((((......(((((....)))))....))))).).......------- ( -30.70, z-score = -1.17, R) >droYak2.chrX 15924436 115 + 21770863 UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGCAGCUCGCCGGCUGAGAAAUGGAUACCGAAUUCCGAAAA ......(((((((.......(((((((((.....)))).(((((......))))).)))))((((........)))).....).)))))).....((((........)))).... ( -31.90, z-score = -1.79, R) >droSec1.super_17 1151728 108 + 1527944 UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCAAUAAAACGCAGCUCGCCGGCUGAGAAAUGGGUCCCGAAAA------- ......(((((((.......(((((((((.....)))).(((((......))))).)))))((((........)))).....).))))))..................------- ( -28.70, z-score = -0.57, R) >droSim1.chrX 3432303 108 - 17042790 UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCAAUAAAACGCAGCUCGCCGGCUGAGAAAUGGGUCCCGAAAA------- ......(((((((.......(((((((((.....)))).(((((......))))).)))))((((........)))).....).))))))..................------- ( -28.70, z-score = -0.57, R) >droPer1.super_18 475155 107 + 1952607 UAAUGACAGCCGCAUCAAUCCAGUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGUAACCCCAGCGAUGAGAAAUGGCCCCAAAAA-------- ....(((.(((((...........(((((.....)))))(((((......)))))...))))))))..................((.((.....)).))........-------- ( -22.40, z-score = -1.49, R) >dp4.chrXL_group1e 1634281 107 + 12523060 UAAUGACAGCCGCAUCAAUCCAGUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGUAACCCCAGCGAUGAGAAAUGGCCCCAAAAA-------- ....(((.(((((...........(((((.....)))))(((((......)))))...))))))))..................((.((.....)).))........-------- ( -22.40, z-score = -1.49, R) >droMoj3.scaffold_6473 1503683 103 - 16943266 UAAUGACAGCUGCAUCAAUCCUGUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGGCGACGUCGACGCGGCAGUCAUAAAACGAGCUGCGACUG------------ ..(((((.(((((.......(((.(((((.....)))))(((((......))))).)))(((((....)))))..))))).))))).................------------ ( -30.80, z-score = -0.56, R) >consensus UAAUGACAGCCGCAUCAAUCCGCUGCGCUAACAAAGCGUUGAAGAAAUCACUUUACCAGCGGCGUCCCAUAAAACGCAGCUCGCCGGCUGAGAAAUGGGUCCCGAAA________ ....(((.(((((...........(((((.....)))))(((((......)))))...))))))))................................................. (-19.43 = -19.44 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:32 2011