
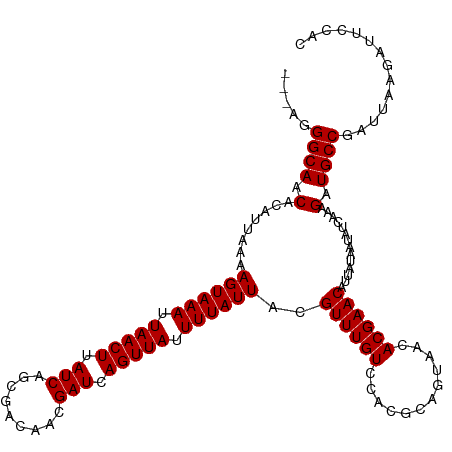
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,289,235 – 17,289,352 |
| Length | 117 |
| Max. P | 0.805006 |

| Location | 17,289,235 – 17,289,352 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.20 |
| Shannon entropy | 0.08136 |
| G+C content | 0.34498 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17289235 117 + 22422827 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droSim1.chrX 3416723 117 - 17042790 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droSec1.super_17 1135926 117 + 1527944 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droEre2.scaffold_4690 7630088 117 + 18748788 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droYak2.chrX 15907973 117 + 21770863 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droAna3.scaffold_13248 1685977 117 - 4840945 ---AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >dp4.chrXL_group1e 1611050 116 + 12523060 ---AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUUCACGCAGCAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCG- ---..((((.(........((((((.(((((.(((.........))).))))).)))))).((((((((.....))))).))).................).)))).............- ( -18.40, z-score = -1.43, R) >droPer1.super_18 452060 116 + 1952607 ---AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGCAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCG- ---..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............- ( -17.90, z-score = -1.24, R) >droWil1.scaffold_180702 442206 118 + 4511350 --UAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --...((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.32, R) >droVir3.scaffold_12970 5721515 118 - 11907090 CGAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCAUGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUUC-- .....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).))))............-- ( -17.90, z-score = -0.72, R) >droMoj3.scaffold_6473 15420453 119 + 16943266 CGAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCAUGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCUC- .....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............- ( -17.90, z-score = -0.58, R) >droGri2.scaffold_14853 9129810 117 + 10151454 CGAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAAAGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUC--- .....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).))))...........--- ( -17.90, z-score = -1.16, R) >consensus ___AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC .....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. (-17.90 = -17.90 + 0.00)


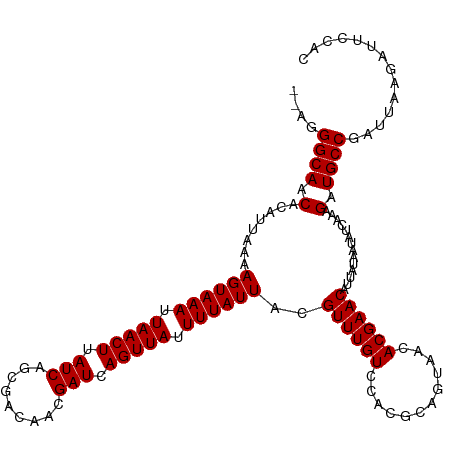
| Location | 17,289,235 – 17,289,352 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.48 |
| Shannon entropy | 0.07776 |
| G+C content | 0.34333 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.805006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17289235 117 + 22422827 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droSim1.chrX 3416723 117 - 17042790 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droSec1.super_17 1135926 117 + 1527944 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droEre2.scaffold_4690 7630088 117 + 18748788 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droYak2.chrX 15907973 117 + 21770863 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >droAna3.scaffold_13248 1685977 117 - 4840945 --AGGGCAGCACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.01, R) >dp4.chrXL_group1e 1611050 116 + 12523060 --AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUUCACGCAGCAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCG- --..((((.(........((((((.(((((.(((.........))).))))).)))))).((((((((.....))))).))).................).)))).............- ( -18.40, z-score = -1.43, R) >droPer1.super_18 452060 116 + 1952607 --AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGCAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCG- --..((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............- ( -17.90, z-score = -1.24, R) >droWil1.scaffold_180702 442206 118 + 4511350 -UAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC -...((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -1.32, R) >droVir3.scaffold_12970 5721516 117 - 11907090 GAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCAUGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUUC-- ....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).))))............-- ( -17.90, z-score = -0.98, R) >droMoj3.scaffold_6473 15420454 119 + 16943266 GAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCAUGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCUCA ....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. ( -17.90, z-score = -0.77, R) >droGri2.scaffold_14853 9129811 116 + 10151454 GAAGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAAAGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUC--- ....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).))))...........--- ( -17.90, z-score = -1.40, R) >consensus __AGGGCAACACAUUAAAAGUAAAUUAACUUAUCAGCGACAACGAUCAGUUAUUUUAUUACGUUUGUCCACGCAGUAACACGAACAUUAUAAUAUCAAAGAUGCCGAUUAAGAUUCCAC ....((((.(........((((((.(((((.(((.........))).))))).))))))..((((((............))))))..............).)))).............. (-17.90 = -17.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:29 2011