| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,264,981 – 17,265,087 |
| Length | 106 |
| Max. P | 0.968330 |

| Location | 17,264,981 – 17,265,087 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.59774 |
| G+C content | 0.45557 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -15.82 |
| Energy contribution | -17.71 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
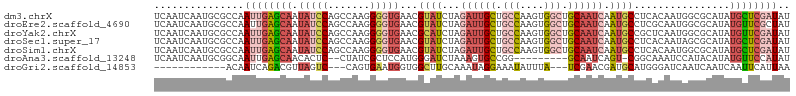
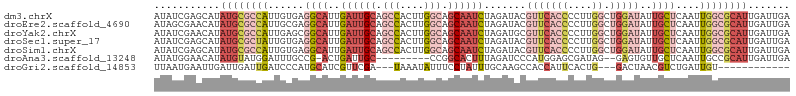
>dm3.chrX 17264981 106 + 22422827 UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGUAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCUCACAAUGGCGCAUAUGCUCGAUAU .......((((((((..((((((.(((((.......)))))..........((((((.(((....))).))))))..)).))))...))))))))........... ( -36.50, z-score = -2.29, R) >droEre2.scaffold_4690 7606587 106 + 18748788 UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGUAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCUCGCAAUGGCGCAUAUGUUCGCUAU .........(((((..(((.((........)))))..)))(((((((((..((((((.(((....))).))))))...(((.......)))).))))))))))... ( -37.00, z-score = -2.08, R) >droYak2.chrX 15881795 106 + 21770863 UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGCAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCGCUCAAUGGCGCAUAUGUUCGAUAU .......((((((((.((((((..(((((.......)))))...((((...((((((.(((....))).)))))).)))).))))))))))))))........... ( -43.90, z-score = -4.05, R) >droSec1.super_17 1111638 106 + 1527944 UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGUAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCUCACAAUAGCGCAUAUGCUCGAUAU .......(((((((..(((.((........)))))..)((((..((((...((((((.(((....))).)))))).)))).))))....))))))........... ( -31.90, z-score = -1.38, R) >droSim1.chrX 3394475 106 - 17042790 UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGUAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCUCACAAUGGCGCAUAUGCUCGAUAU .......((((((((..((((((.(((((.......)))))..........((((((.(((....))).))))))..)).))))...))))))))........... ( -36.50, z-score = -2.29, R) >droAna3.scaffold_13248 1656646 94 - 4840945 UCAAUCAAUGCGGCAAUUGAGCAACACUC--CUAUCGCUCCAUGGGAUCUAAAGUGCCGG---------GCAAUCAGU-CGGCAAAUCCAUACAUAUGUUCCAUAU .......(((..(((...((((.......--.....))))....((((......((((((---------........)-))))).)))).......)))..))).. ( -18.60, z-score = 0.30, R) >droGri2.scaffold_14853 9099177 88 + 10151454 ------------ACAAUCAGACGUUAGUC---CAGUGAAUGGUGGCUUGCAAAUAGGAAAUAUUUA---UCGAACGAUGCAUGGGAUCAAUCAAUCAAUUCAUUAA ------------.......(((....)))---...(((.((((..(.((((((((.....)))))(---((....)))))).)..)))).)))............. ( -11.40, z-score = 1.58, R) >consensus UCAAUCAAUGCGCCAAUUGAGCAAUAUCCAGCCAAGGGGUGAACGUAUCUAGAUUGCUGCCAAGUGGCUGCAAUCAAUGCCUCACAAUGGCGCAUAUGUUCGAUAU ...............((((((((.(((((.......)))))...((((...((((((.(((....))).)))))).))))................)))))))).. (-15.82 = -17.71 + 1.90)
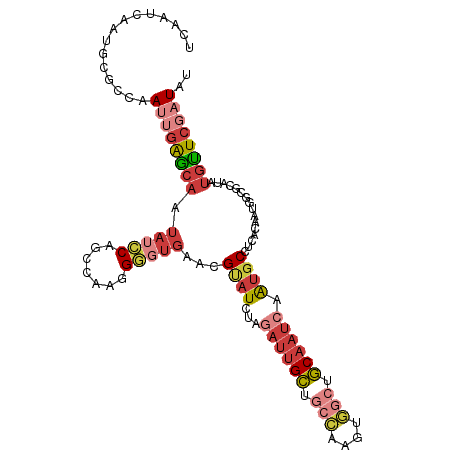
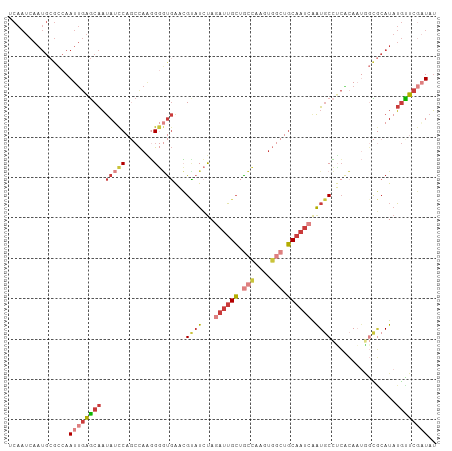
| Location | 17,264,981 – 17,265,087 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.59774 |
| G+C content | 0.45557 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -15.17 |
| Energy contribution | -16.94 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
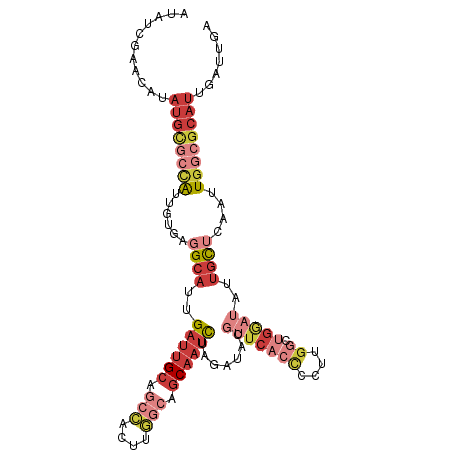
>dm3.chrX 17264981 106 - 22422827 AUAUCGAGCAUAUGCGCCAUUGUGAGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUACGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ...((((.((.((((((((...((((.((..((((((.(((....))).)))))).......(((((((....)).)))))..))))))..)))))))))).)))) ( -39.50, z-score = -2.71, R) >droEre2.scaffold_4690 7606587 106 - 18748788 AUAGCGAACAUAUGCGCCAUUGCGAGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUACGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ....(((.((.(((((((((((...((((..((((((.(((....))).)))))).......(((((((....)).)))))..))))))).)))))))))).))). ( -36.40, z-score = -1.92, R) >droYak2.chrX 15881795 106 - 21770863 AUAUCGAACAUAUGCGCCAUUGAGCGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUGCGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ...((((.((.((((((((((((((((((((((((((.(((....))).))))))..)))))(((((((....)).)))))..))))))).)))))))))).)))) ( -48.00, z-score = -5.05, R) >droSec1.super_17 1111638 106 - 1527944 AUAUCGAGCAUAUGCGCUAUUGUGAGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUACGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ...((((.((.((((((((...((((.((..((((((.(((....))).)))))).......(((((((....)).)))))..))))))..)))))))))).)))) ( -37.20, z-score = -2.18, R) >droSim1.chrX 3394475 106 + 17042790 AUAUCGAGCAUAUGCGCCAUUGUGAGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUACGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ...((((.((.((((((((...((((.((..((((((.(((....))).)))))).......(((((((....)).)))))..))))))..)))))))))).)))) ( -39.50, z-score = -2.71, R) >droAna3.scaffold_13248 1656646 94 + 4840945 AUAUGGAACAUAUGUAUGGAUUUGCCG-ACUGAUUGC---------CCGGCACUUUAGAUCCCAUGGAGCGAUAG--GAGUGUUGCUCAAUUGCCGCAUUGAUUGA ....((....((((...((((((((((-.........---------.))))).....)))))))))((((((((.--...)))))))).....))........... ( -21.90, z-score = 0.32, R) >droGri2.scaffold_14853 9099177 88 - 10151454 UUAAUGAAUUGAUUGAUUGAUCCCAUGCAUCGUUCGA---UAAAUAUUUCCUAUUUGCAAGCCACCAUUCACUG---GACUAACGUCUGAUUGU------------ ....(((((.((((((.((((.......)))).))))---)(((((.....)))))........).)))))..(---(((....))))......------------ ( -11.10, z-score = 0.83, R) >consensus AUAUCGAACAUAUGCGCCAUUGUGAGGCAUUGAUUGCAGCCACUUGGCAGCAAUCUAGAUACGUUCACCCCUUGGCUGGAUAUUGCUCAAUUGGCGCAUUGAUUGA ...........((((((((............((((((.(((....))).))))))..................(((........)))....))))))))....... (-15.17 = -16.94 + 1.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:25 2011