
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,679,650 – 11,679,781 |
| Length | 131 |
| Max. P | 0.670248 |

| Location | 11,679,650 – 11,679,781 |
|---|---|
| Length | 131 |
| Sequences | 6 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 83.37 |
| Shannon entropy | 0.30860 |
| G+C content | 0.47128 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -19.99 |
| Energy contribution | -21.77 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
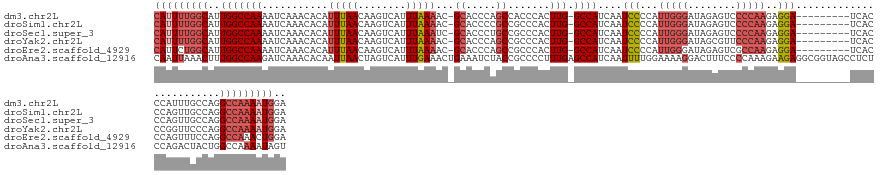
>dm3.chr2L 11679650 131 + 23011544 CAUUUUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAAC-GCACCCAGCCACCCACUUG-GCCAUCAAUCCCCAUUGGGAUAGAGUCCCCAAGAGGA---------UCACCCAUUUGCCAGGCCAAAAUGGA (((((((((.(((((..................................-((.....))...((.((((-(...((.(((((....))))).))....))))).)).---------..........)))))))))))))).. ( -37.30, z-score = -2.81, R) >droSim1.chr2L 11489117 131 + 22036055 CAUUUUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAAC-GCACCCCGCCGCCCACUUG-GCCAUCAAUCCCCAUUGGGAUAGAGUCCCCAAGAGGA---------UCACCCAGUUGCCAGGCCAAAAUGGA (((((((((.(((((..................................-((.....))...((.((((-(...((.(((((....))))).))....))))).)).---------..........)))))))))))))).. ( -37.30, z-score = -2.22, R) >droSec1.super_3 7071585 131 + 7220098 CAUUUUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAUC-GCACCCUGCCGCCCACUUG-GCCAUCAAUCCCCAUUGGGAUAGAGUCCCCAAGAGGA---------UCACCCAGUUGCCAGGCCAAAAUGGA (((((((((.(((((..................................-((.....))...((.((((-(...((.(((((....))))).))....))))).)).---------..........)))))))))))))).. ( -37.60, z-score = -2.00, R) >droYak2.chr2L 8102470 131 + 22324452 CAUUUUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAAC-GCACCCAGCCGCCCACUUG-GCCAUCAAUCCCCAUUGGGAUAGCGUUCCCAAGAGGA---------UCACCCGGUUCCCAGGCCAAAAUGGA (((((((((..(((((((...........(((((........)))))..-((.....)).......)))-))))...((((...((((((......))))))..)))---------)..............))))))))).. ( -36.40, z-score = -1.76, R) >droEre2.scaffold_4929 12910569 131 - 26641161 CAUUCUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAAC-GCACCCAGCCGCCCACUUG-GCCAUCAAUCCCCAUUGGGAUAGAGUCGCCAAGAGGA---------UCACCCAGUUUCCAGGCCAAACUGGA ...((..(..((((((.................................-((.....))...((.((((-((..((.(((((....))))).))...)))))).)).---------..............)))))).)..)) ( -34.90, z-score = -1.82, R) >droAna3.scaffold_12916 9397040 142 - 16180835 CAAUUAAACUUUGGCCAAGAUCAAACACAAUUAACUAGUCAUUUGAAACUGAAAUCUACCGCCCCUUUGAGCCAUCAAUUUUGGAAAAGGACUUUCCCCAAAGAAGAGGCGGUAGCCUCUCCAGACUACUGGCCAAAAUAGU .........((((((((...(((((.((.........))..)))))..(((....(((((((((((((...(((.......))).))))).((((.......)))).))))))))......))).....))))))))..... ( -37.60, z-score = -2.93, R) >consensus CAUUUUGGCAUUGGCCAAAAUCAAACACAUUUAACAAGUCAUUUAAAAC_GCACCCAGCCGCCCACUUG_GCCAUCAAUCCCCAUUGGGAUAGAGUCCCCAAGAGGA_________UCACCCAGUUGCCAGGCCAAAAUGGA (((((((((..(((((((...........(((((........)))))...((.....)).......))).))))......((..(((((........)))))..)).........................))))))))).. (-19.99 = -21.77 + 1.78)


| Location | 11,679,650 – 11,679,781 |
|---|---|
| Length | 131 |
| Sequences | 6 |
| Columns | 142 |
| Reading direction | reverse |
| Mean pairwise identity | 83.37 |
| Shannon entropy | 0.30860 |
| G+C content | 0.47128 |
| Mean single sequence MFE | -46.14 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
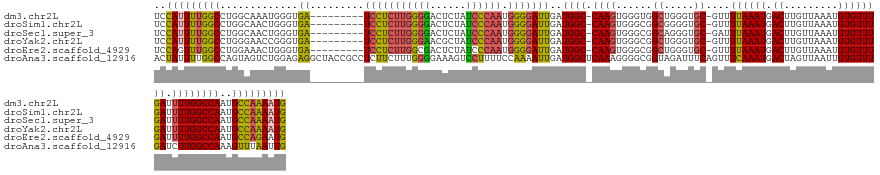
>dm3.chr2L 11679650 131 - 23011544 UCCAUUUUGGCCUGGCAAAUGGGUGA---------UCCUCUUGGGGACUCUAUCCCAAUGGGGAUUGAUGGC-CAAGUGGGUGGCUGGGUGC-GUUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAAAAUG ..(((((((((.((((.....(((.(---------(((.(((((...(((.(((((....))))).)).).)-)))).)))).)))(((..(-((((.............)))))..))).......))))..))))))))) ( -45.82, z-score = -1.87, R) >droSim1.chr2L 11489117 131 - 22036055 UCCAUUUUGGCCUGGCAACUGGGUGA---------UCCUCUUGGGGACUCUAUCCCAAUGGGGAUUGAUGGC-CAAGUGGGCGGCGGGGUGC-GUUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAAAAUG (((((((.((((.(....).....((---------(((((((((((......)))))).)))))))...)))-)))))))).((((((...(-((.....))).))))))...........((((((((....)))))))). ( -44.00, z-score = -0.88, R) >droSec1.super_3 7071585 131 - 7220098 UCCAUUUUGGCCUGGCAACUGGGUGA---------UCCUCUUGGGGACUCUAUCCCAAUGGGGAUUGAUGGC-CAAGUGGGCGGCAGGGUGC-GAUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAAAAUG ..((((((((((((....).))..((---------(((((((((((......)))))).)))))))..((((-(((.(..(((.((....((-((.((....)).))))....)))))..)...)))))))..))))))))) ( -44.10, z-score = -0.98, R) >droYak2.chr2L 8102470 131 - 22324452 UCCAUUUUGGCCUGGGAACCGGGUGA---------UCCUCUUGGGAACGCUAUCCCAAUGGGGAUUGAUGGC-CAAGUGGGCGGCUGGGUGC-GUUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAAAAUG ..(((((((((((((...))))..((---------(((((((((((......)))))).)))))))..((((-(((.(..(((((.....))-(((......)))..........)))..)...)))))))..))))))))) ( -46.80, z-score = -1.73, R) >droEre2.scaffold_4929 12910569 131 + 26641161 UCCAGUUUGGCCUGGAAACUGGGUGA---------UCCUCUUGGCGACUCUAUCCCAAUGGGGAUUGAUGGC-CAAGUGGGCGGCUGGGUGC-GUUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAGAAUG .((((((..(((((....).))))(.---------(((.((((((.(.((.(((((....))))).))).))-)))).))))))))))(..(-((((.............)))))..)...((((((((....)))))))). ( -46.52, z-score = -2.07, R) >droAna3.scaffold_12916 9397040 142 + 16180835 ACUAUUUUGGCCAGUAGUCUGGAGAGGCUACCGCCUCUUCUUUGGGGAAAGUCCUUUUCCAAAAUUGAUGGCUCAAAGGGGCGGUAGAUUUCAGUUUCAAAUGACUAGUUAAUUGUGUUUGAUCUUGGCCAAAGUUUAAUUG ...(((((((((((....((((((...(((((((((((..(((((..((.....))..))))).((((....))))))))))))))).))))))..((((((.((.........))))))))..)))))))))))....... ( -49.60, z-score = -3.49, R) >consensus UCCAUUUUGGCCUGGCAACUGGGUGA_________UCCUCUUGGGGACUCUAUCCCAAUGGGGAUUGAUGGC_CAAGUGGGCGGCUGGGUGC_GUUUUAAAUGACUUGUUAAAUGUGUUUGAUUUUGGCCAAUGCCAAAAUG ..(((((((((........................(((((((((((......)))))).)))))....((((.((((..(((.((.....)).)))((((((.((.........)))))))).))))))))..))))))))) (-27.74 = -28.08 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:17 2011