| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,263,291 – 17,263,410 |
| Length | 119 |
| Max. P | 0.710703 |

| Location | 17,263,291 – 17,263,410 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.57482 |
| G+C content | 0.40688 |
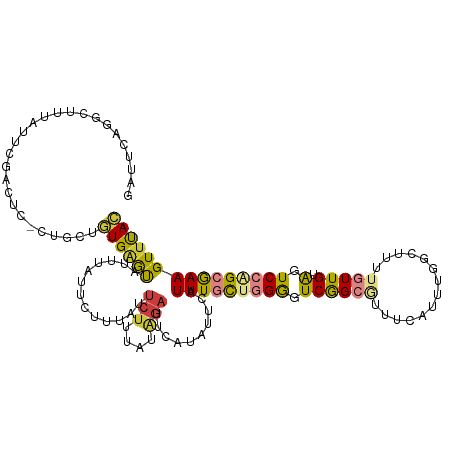
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -10.56 |
| Energy contribution | -10.55 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
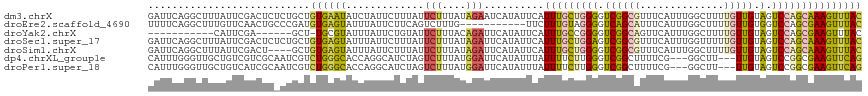
>dm3.chrX 17263291 119 - 22422827 GAUUCAGGCUUUAUUCGACUCUCUGCUGUGAAUAUCUAUUCUUUAUUCUUUAUAGAAUCAUAUUCAUUUGCUGGGGUCGGCGUUUCAUUUGGCUUUUGUUGUAGUCCAGCAAAGUUUAC .....(((((((..((((((((..((.((((((((..(((((.((.....)).))))).))))))))..)).))))))))........((((((........)).)))).))))))).. ( -29.10, z-score = -2.63, R) >droEre2.scaffold_4690 7604964 108 - 18748788 UUUUCAGGCUUUGUUCAACUGCCCGAUGUGAGUAUUUAUUCUUCAGUCUUUG-----------UUCUUUGUAGGGGUCAGCAUUUCAUUUGGCUUUUGUUGUGGUCCAGCGAAGUUUAC .....((((((((((..((..(..(((..((((....))))....)))....-----------.......((((((((((........))))))))))..)..))..)))))))))).. ( -22.40, z-score = -0.26, R) >droYak2.chrX 15879803 101 - 21770863 -----------CAUUCGA------GCU-UGCGUAUUUAUUCUGUAUUCUUUACAGAUUCAUAUUCAUUUGCCGGGGUCGGCAGUUCAUUUGGCUUUUGUUGUAGUCCAGCGAAGUUUAC -----------.....((------((.-((.((((....((((((.....))))))...)))).))..(((((....)))))))))....((((((.((((.....))))))))))... ( -24.20, z-score = -2.01, R) >droSec1.super_17 1109940 119 - 1527944 GAUUCAGGCUUUAUUCGACUCUCUGCUGUGAGUAUUUAUUCUUUAUUCUUUAUAGAUUCAUAUUCAUUUGCUGGAGUCGGCGUUUCAUUUGGUUUUUGUUGUAGUCCAGCAAAGUUUAC .....(((((((..((((((((..((.((((((((....(((.((.....)).)))...))))))))..)).)))))))).................((((.....))))))))))).. ( -27.50, z-score = -2.40, R) >droSim1.chrX 3392772 115 + 17042790 GAUUCAGGCUUUAUUCGACU----GCUGUGAGUAUUUAUUCUUUAUUCUUUAUAGAUUCAUAUUCAUUUGCUGGGGUCGGCGUUUCAUUUGGCUUUUGUUGUAGUCCAGCAAAGUUUAC .....(((((((.((.((((----((.((((((((....(((.((.....)).)))...))))))))..((..(((((((........)))))))..)).)))))).)).))))))).. ( -24.30, z-score = -1.12, R) >dp4.chrXL_group1e 1579741 113 - 12523060 CAUUUGGGUUGCUGUCGUCGCAAUCGUCUGGGCACCAGGCAUCUAGUCUUUAUGGAUUCAUAUUUAUUUUCUUGGGUCGGCUUUUCG---GGCUU---UUGUAGUCCGGCGAAGUUCAG ......((((((.......))))))((((((...))))))..............((((((............))))))(((((((((---((((.---....))))))).))))))... ( -30.70, z-score = -1.56, R) >droPer1.super_18 421796 113 - 1952607 CAUUUGGGUUGCUGUCAUCGCAAUCGUCUGGGCACCAGGCAUCUAGUCUUUAUGGAUUCAUAUUUAUUUUCUUGGGUCGGCUUUUCG---GGCUU---UUGUAGUCCGGCGAAGUUCAG ......((((((.......))))))((((((...))))))..............((((((............))))))(((((((((---((((.---....))))))).))))))... ( -30.70, z-score = -1.74, R) >consensus GAUUCAGGCUUUAUUCGACUC_CUGCUGUGAGUAUUUAUUCUUUAUUCUUUAUAGAUUCAUAUUCAUUUGCUGGGGUCGGCGUUUCAUUUGGCUUUUGUUGUAGUCCAGCGAAGUUUAC ...........................((((((.............(((....)))..........(((((((((.((((((..............))))).).))))))))))))))) (-10.56 = -10.55 + -0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:24 2011