
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,248,588 – 17,248,686 |
| Length | 98 |
| Max. P | 0.627138 |

| Location | 17,248,588 – 17,248,686 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Shannon entropy | 0.40555 |
| G+C content | 0.45479 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.21 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
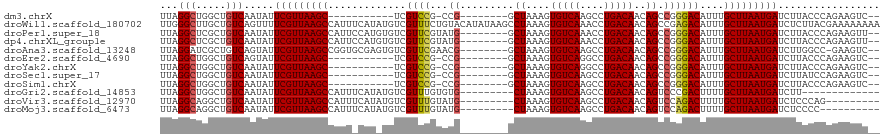
>dm3.chrX 17248588 98 - 22422827 UUAGGCUGGCUGUCAAUAUUCGUUAAGC-----------UCGUCCG-CCG--------GCUAAAGUGUCAAGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUC-- ...((((..(((.......(((((((((-----------..((((.-.((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -34.64, z-score = -3.34, R) >droWil1.scaffold_180702 380874 120 - 4511350 UUGGGCUUGCUGUCAGUUUUCGUUAAGCCAUUUCAUAUGUCGUUUCUGUACAUAUAAGCCUAAAGUGUCAAACCUGACAACAGCCGAGACAUUUGCUUAAUGAUCUCUUACGAAAAAAAA (((((((((((....(....)....)))......((((((((....)).))))))))))))))..(((((....)))))......((((((((.....)))).))))............. ( -24.50, z-score = -0.84, R) >droPer1.super_18 400956 110 - 1952607 UUAGGCUCGCUGUCAAUAUUCGUUAAGCCAUUCCAUGUGUCGUUCGUAUG--------GCUAAAGUGUCAAACCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUU-- ...((((..(((.......((((((((((((.....)))..((((...((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -27.34, z-score = -1.19, R) >dp4.chrXL_group1e 1558762 110 - 12523060 UUAGGCUCGCUGUCAAUAUUCGUUAAGCCAUUCCAUGUGUCGUUCGUAUG--------GCUAAAGUGUCAAACCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUU-- ...((((..(((.......((((((((((((.....)))..((((...((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -27.34, z-score = -1.19, R) >droAna3.scaffold_13248 1633514 109 + 4840945 UUAGGAUCGCUGUCAGUAUUCGUUAAGCCGGUGCGAGUGUCGUUCGAACG--------GCUAAAGUGUCAAGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUGGCC-GAAGUC-- ....(((..(.(((((...(((((((((..(((((((.....))))..((--------(((....(((((....)))))..)))))...)))..)))))))))..))))).-)..)))-- ( -35.00, z-score = -1.31, R) >droEre2.scaffold_4690 7590805 98 - 18748788 UUAGGCUGGCUGUCAGUAUUCGUUAAGC-----------UCGUCCG-CCG--------GCUAAAGUGUCAGGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUC-- ...((((..(((...(((.(((((((((-----------..((((.-.((--------(((....(((((....)))))..)))))))))....)))))))))...))).))).))))-- ( -36.30, z-score = -3.07, R) >droYak2.chrX 15864836 98 - 21770863 UUAGGCUGGCUGUCAAUAUUCGUUAAGC-----------UCGUCCG-CCG--------GCUAAAGUGUCAGGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUC-- ...((((..(((.......(((((((((-----------..((((.-.((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -35.34, z-score = -3.11, R) >droSec1.super_17 1094413 98 - 1527944 UUAGGCUGGCUGUCAAUAUUCGUUAAGC-----------UCGUCCG-CCG--------GCUAAAGUGUCAAGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUAUCCAGAAGUC-- ...((((..(((.......(((((((((-----------..((((.-.((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -34.64, z-score = -3.49, R) >droSim1.chrX 3378983 98 + 17042790 UUAGGCUGGCUGUCAAUAUUCGUUAAGC-----------UCGUCCG-CCG--------GCUAAAGUGUCAAGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUC-- ...((((..(((.......(((((((((-----------..((((.-.((--------(((....(((((....)))))..)))))))))....))))))))).......))).))))-- ( -34.64, z-score = -3.34, R) >droGri2.scaffold_14853 9076062 98 - 10151454 UUAGGCUGGCUGUCAAUAUUCGUUAAGCCAUUUCAUAUGUCGUUUGUGUG---------CUAAAGUGUCAAGCCUGACAACAGUCCCGACUUUUGCUUAAUGAUCUU------------- ...(((.....))).....(((((((((.........(((((((((.(..---------(....)..))))))..))))..(((....)))...)))))))))....------------- ( -20.30, z-score = -0.35, R) >droVir3.scaffold_12970 5670276 102 + 11907090 UUAGGCAGGCUGUCAAUAUUCGUUAAGCCAUUUCAUAUGUCGUUUGUAUG---------CUAAAGUGUCAAGCCUGACAACAGUCCAGACUUUUGCUUAAUGAUCUCCCAG--------- (((((((((((..............)))).........((((((((.(((---------(....)))))))))..(((....)))..)))...)))))))...........--------- ( -21.54, z-score = -0.84, R) >droMoj3.scaffold_6473 15369179 101 - 16943266 UUAGGCAGGCUGUCAAUAUUCGUUAAGCCAUUUCAUAUGUCGUUUGUAUG---------CUAAAGUGUCAAGCCUGACAACAGUCCAGACUUUUGCUUAAUGAUCUCCCC---------- (((((((((((..............)))).........((((((((.(((---------(....)))))))))..(((....)))..)))...)))))))..........---------- ( -21.54, z-score = -1.04, R) >consensus UUAGGCUGGCUGUCAAUAUUCGUUAAGCCAUU_CAU_UGUCGUUCGUACG________GCUAAAGUGUCAAGCCUGACAACAGCCGGGACAUUUGCUUAAUGAUCUUACCCAGAAGUC__ ...(((.....))).....((((((((((((.....)))..((((..............(....)(((((....))))).......))))....)))))))))................. (-14.32 = -14.21 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:21 2011