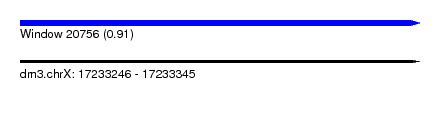
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,233,246 – 17,233,345 |
| Length | 99 |
| Max. P | 0.907799 |

| Location | 17,233,246 – 17,233,345 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Shannon entropy | 0.15685 |
| G+C content | 0.34375 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -18.77 |
| Energy contribution | -18.77 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907799 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 17233246 99 + 22422827 -CACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCUCUCG- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))..................- ( -19.02, z-score = -1.39, R) >droSim1.chrX 3363905 100 - 17042790 -CACCAUUAUGUAACAAAAUUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCUCUCG- -.....((((((((((((.....)))).....((((((.(((((.............)))))....))))))...))))))))..................- ( -19.22, z-score = -1.49, R) >droSec1.super_17 1079390 99 + 1527944 -CACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCUCUCG- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))..................- ( -19.02, z-score = -1.39, R) >droYak2.chrX 15849432 97 + 21770863 -CACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCCCU--- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))................--- ( -19.02, z-score = -1.39, R) >droEre2.scaffold_4690 7572831 97 + 18748788 -CACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCUCU--- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))................--- ( -19.02, z-score = -1.32, R) >dp4.chrXL_group1e 1539793 100 + 12523060 -UACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGGCUCUCUCUUUC -..(((((((((((((((-....)))).....((((((.(((((.............)))))....))))))...)))))))).....)))........... ( -19.72, z-score = -1.32, R) >droPer1.super_18 381974 100 + 1952607 -UACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGGCUCUCUCUUUC -..(((((((((((((((-....)))).....((((((.(((((.............)))))....))))))...)))))))).....)))........... ( -19.72, z-score = -1.32, R) >droWil1.scaffold_180702 4150768 91 - 4511350 UAACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUU---GCCUC------- ......((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))....---.....------- ( -19.02, z-score = -1.27, R) >droVir3.scaffold_12970 5652806 90 - 11907090 -CUCCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUU---GCCCC------- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))....---.....------- ( -19.02, z-score = -1.52, R) >droGri2.scaffold_14853 9056251 90 + 10151454 -CACCAUUAUGUAACAAA-UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUU--GCCUC-------- -.....((((((((((((-....)))).....((((((.(((((.............)))))....))))))...))))))))....--.....-------- ( -19.02, z-score = -1.21, R) >consensus _CACCAUUAUGUAACAAA_UUACUUUGGCAAAGUGCUCGUGUAUUCGCUUUUUAAAUAUGCAAAAAGAGCGCAAUUUGCAUAAAUUUUUGCCUCUCUCU___ ......((((((((((((.....)))).....((((((.(((((.............)))))....))))))...))))))))................... (-18.77 = -18.77 + -0.00)

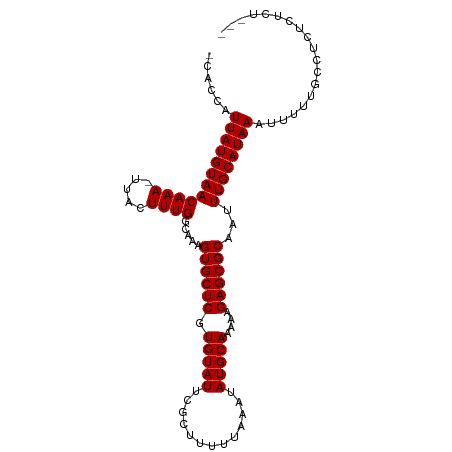
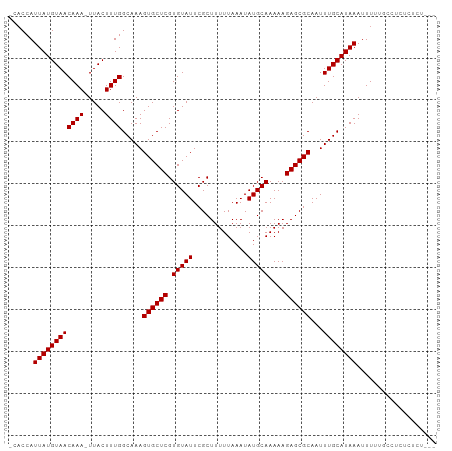
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:17 2011