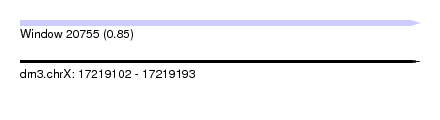
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,219,102 – 17,219,193 |
| Length | 91 |
| Max. P | 0.847280 |

| Location | 17,219,102 – 17,219,193 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.07 |
| Shannon entropy | 0.59209 |
| G+C content | 0.50744 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -10.73 |
| Energy contribution | -10.41 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17219102 91 + 22422827 -------UUAAUUUAUGGGGUUGU--GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGG----------CACACACACACAACCCCUGGCCAAGUAUCCCUGCUCCU -------.........((((((((--(.....((((((...))))))((..(......)..)----------).......))))))))).((...((((....)))))). ( -28.10, z-score = -1.53, R) >droWil1.scaffold_180702 339457 88 + 4511350 -------UUAAUUUAUGGGGUUGCCUGGCUAAGCGAAUGCAAUUUGCUCUGCCCGCUGUCGGCAA------UCGAACUACCGGGGCCAAAGACCAACCCCU--------- -------.........(((((((...(((..(((((((...)))))))..)))....((((((..------(((......))).)))...)))))))))).--------- ( -29.60, z-score = -1.31, R) >dp4.chrXL_group1e 1522903 80 + 12523060 -------CUAAUUUAUGGGGUUGC--GGCUAAGCGAAUUCAAUUUGCGUUGCCUUCGAGUGGCGGG-----CCGCAGCCGCACAACCCCUA--CAA-------------- -------.........((((((((--((((..((((((...))))))((.((((.(.....).)))-----).)))))))))..)))))..--...-------------- ( -33.00, z-score = -2.46, R) >droAna3.scaffold_13248 1594552 70 - 4840945 -------UUAAUUUAUGGGGUUGC--GGCUAAGCGAAUUCAAUUUGCGUUGCCUUCGAGUG----------CCGCACACACACACCCCU--------------------- -------.........((((((((--(((....((((..((((....))))..))))...)----------))))).......))))).--------------------- ( -24.41, z-score = -2.87, R) >droEre2.scaffold_4690 7559405 89 + 18748788 -------UUAAUUUAUGGGGUUGC--GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGG----------CACACACACA--ACCCCUUGCCAAGUAUCCGAACUCCU -------.........(((((((.--(((...((((((...))))))...))).....(((.----------....))).))--)))))..................... ( -25.50, z-score = -1.56, R) >droYak2.chrX 15834939 78 + 21770863 -------UUAAUUUAUGGGGUUGC--GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGGCAC------ACACACAAGUAGCACCACUCCU----------------- -------.........((((((((--(((...((((((...))))))...))).....(((....------.))).......)))..))))).----------------- ( -22.20, z-score = -1.25, R) >droSec1.super_17 1065340 89 + 1527944 -------UUAAUUUAUGGGGUUGU--GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGG----------CACACACACA--ACCCCUGGCCAAGUAUCCCUGCUCCU -------.........((((((((--(.....((((((...))))))((..(......)..)----------).....))))--))))).((...((((....)))))). ( -28.10, z-score = -1.62, R) >droSim1.chrX 3349632 89 - 17042790 -------UUAAUUUAUGGGGUUGU--GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGG----------CACACACACA--ACCCCUGGCCAAGUAUCCCUGCUCCU -------.........((((((((--(.....((((((...))))))((..(......)..)----------).....))))--))))).((...((((....)))))). ( -28.10, z-score = -1.62, R) >droGri2.scaffold_14853 9037085 97 + 10151454 UCGGCUCUUAAUUUAUGGGGUUGC--GGCUAAGCGAAUGCAAUUUGCGCUGCCUCUCAACGCGCUCAACCCCCAAACAGUUAGAGUCAAUUCACAAACA----------- ..((((((........(((((((.--(((...((....)).....(((.((.....)).))))))))))))).........))))))............----------- ( -28.43, z-score = -2.00, R) >consensus _______UUAAUUUAUGGGGUUGC__GGCUAAGCGAAUUCAAUUUGCGCUGCCUUCGAGUGG__________CACACACACA_AACCCCUGGCCAAGUA___________ .................((.......(((...((((((...))))))...))).....(((...........)))..........))....................... (-10.73 = -10.41 + -0.32)

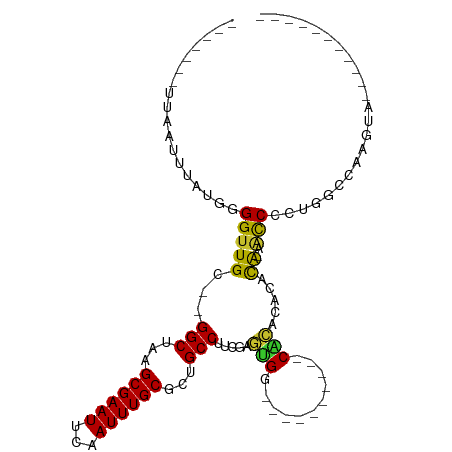
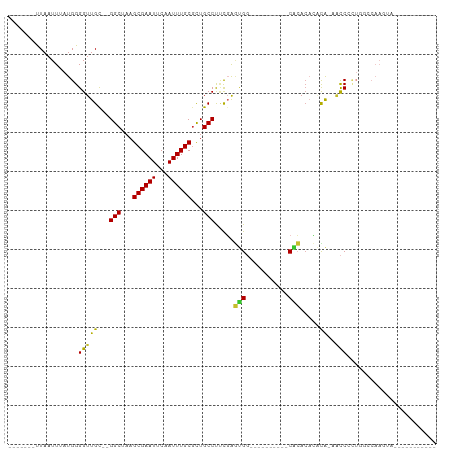
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:16 2011