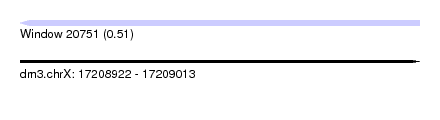
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,208,922 – 17,209,013 |
| Length | 91 |
| Max. P | 0.512704 |

| Location | 17,208,922 – 17,209,013 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.52588 |
| G+C content | 0.67580 |
| Mean single sequence MFE | -42.32 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.34 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.512704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
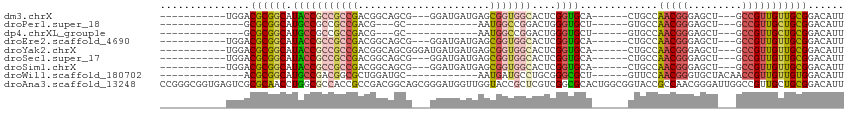
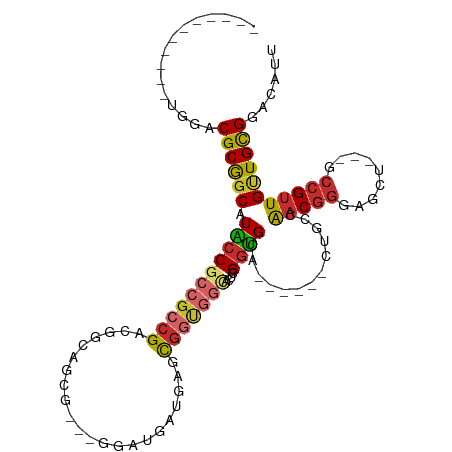
>dm3.chrX 17208922 91 - 22422827 -----------UGGACGCGGCAUACCGCCGCCGACGGCAGCG---GGAUGAUGAGCGGUGGCACUCGGUGCA------CUGCCAACGGGAGCU---GCCGUUGUUGCGGACAUU -----------((..((((((.....)))..((((((((((.---...((.((.((((((.(((...)))))------)))))).))...)))---)))))))..)))..)).. ( -44.90, z-score = -2.40, R) >droPer1.super_18 354517 76 - 1952607 --------------GCGCGGCAUGCCGCCGCCGACG---GC------------AAUGGCCGGACUGGGUGCU------GUGCCAACGGGAGCU---GCCGUUGCUGCGGACAUU --------------(.((((....)))))((((.((---((------------((((((.((.((((((...------..)))..)))...))---))))))))))))).)... ( -34.20, z-score = 0.10, R) >dp4.chrXL_group1e 1512293 76 - 12523060 --------------GCGCGGCAUGCCGCCGCCGACG---GC------------AAUGGCCGGACUGGGUGCU------GUGCCAACGGGAGCU---GCCGUUGCUGCGGACAUU --------------(.((((....)))))((((.((---((------------((((((.((.((((((...------..)))..)))...))---))))))))))))).)... ( -34.20, z-score = 0.10, R) >droEre2.scaffold_4690 7549225 91 - 18748788 -----------UGGACGCGGCAUACCGCCGCCGACGGCAGCG---GGAUGAUGAGCGGUGGCACUCGGUGCA------CUGCCAACGGGAGCU---GCCGUUGUUGCGGACAUU -----------((..((((((.....)))..((((((((((.---...((.((.((((((.(((...)))))------)))))).))...)))---)))))))..)))..)).. ( -44.90, z-score = -2.40, R) >droYak2.chrX 15824617 94 - 21770863 -----------UGGACGCGGCAUACCGCCGCCGACGGCAGCGGGAUGAUGAUGAGCGGUGGCACUCGGUGCA------CUGCCAACGGGAGCU---GCCGUUGUUGCGGACAUU -----------((..((((((.....)))..((((((((((.......((.((.((((((.(((...)))))------)))))).))...)))---)))))))..)))..)).. ( -43.10, z-score = -1.75, R) >droSec1.super_17 1055352 91 - 1527944 -----------UGGACGCGGCAUACCGCCGCCGACGGCAGCG---GGAUGAUGAGCGGUGGCACUCGGUGCA------CUGCCAACGGGAGCU---GCCGUUGUUGCGGACAUU -----------((..((((((.....)))..((((((((((.---...((.((.((((((.(((...)))))------)))))).))...)))---)))))))..)))..)).. ( -44.90, z-score = -2.40, R) >droSim1.chrX 3339388 91 + 17042790 -----------UGGACGCGGCAUACCGCCGCCGACGGCAGCG---GGAUGAUGAGCGGUGGCACUCGGUGCA------CUGCCAACGGGAGCU---GCCGUUGUUGCGGACAUU -----------((..((((((.....)))..((((((((((.---...((.((.((((((.(((...)))))------)))))).))...)))---)))))))..)))..)).. ( -44.90, z-score = -2.40, R) >droWil1.scaffold_180702 327016 82 - 4511350 --------------ACGCGGCAUGCCGACGGCGCUGGAUGC------------AAUGAUGCCUGCGGGCGCU------GUUCCAACGGGUGCUACAACCGUUGUUGUGGACAUU --------------.((((((.....(((((((((.(..((------------(....)))...).))))))------)))..(((((.........)))))))))))...... ( -30.70, z-score = 0.02, R) >droAna3.scaffold_13248 1584215 114 + 4840945 CCGGGCGGUGAGUCGCGCAACCUGGCGCCACCGCCGACGGCAGCGGGAUGGUUGGUACCGCUCGUCGGCGCACUGGCGGUACCGCCAACGGGAUUGGCCGUUGCUGCGGACAUU ((((((((((....((((......)))))))))))..((((((((........(((((((((.((......)).)))))))))(((((.....))))))))))))))))..... ( -59.10, z-score = -1.29, R) >consensus ___________UGGACGCGGCAUACCGCCGCCGACGGCAGCG___GGAUGAUGAGCGGUGGCACUCGGUGCA______CUGCCAACGGGAGCU___GCCGUUGUUGCGGACAUU ...............((((((.(((((((((((......................)))))))....)))).............(((((.........)))))))))))...... (-19.00 = -19.34 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:13 2011