| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,192,983 – 17,193,083 |
| Length | 100 |
| Max. P | 0.781469 |

| Location | 17,192,983 – 17,193,083 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.59310 |
| G+C content | 0.55997 |
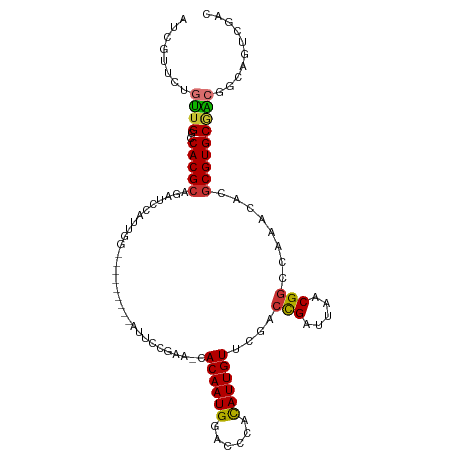
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
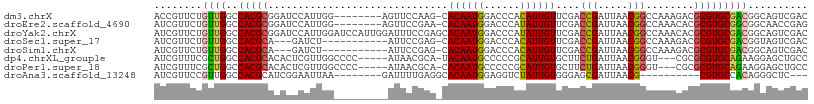
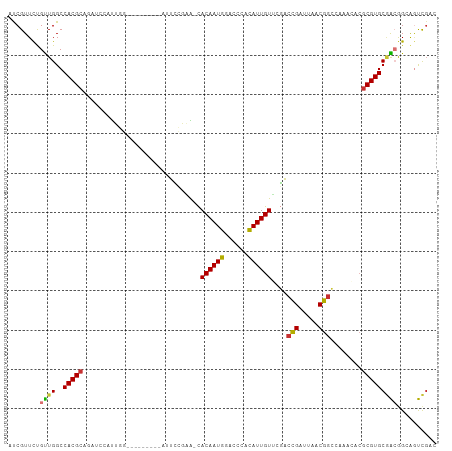
>dm3.chrX 17192983 100 + 22422827 ACCGUUCUGUUGGCCACGCGGAUCCAUUGG--------AGUUCCAAG-CACAAUGGACCCACAUUGUUCGACCGAUUAACGGCCAAAGACGCGUGCGACGGCAGUCGAC ..((..((((((.((((((((.(((((((.--------.((.....)-).))))))).)).........(.(((.....))).)......))))).).)))))).)).. ( -33.40, z-score = -1.03, R) >droEre2.scaffold_4690 7533464 100 + 18748788 AUCGUUCUGUUGGCCACGCGGAUCCAUUGG--------AGUUCCGAA-CACAAUGGACCCAUAUUGUUCGACCGAUUAACGGCCAAACACGCGUGCGGCGGCAACCGAG .(((...(((((.((((((((.(((((((.--------.((.....)-).))))))).))....((((.(.(((.....))).).)))).))))).).)))))..))). ( -35.30, z-score = -1.61, R) >droYak2.chrX 15808023 109 + 21770863 AUCGUUCUGUUGGCCACGCGGAUCCAUUGGAUCCAUUGGAUUUCCGAGCACAAUGGACCCAUAUUGUUCGACCGAUUAACGGCCAAACACGCGUGCGACGGCAGUCGAC .(((..((((((.((((((((((((...)))))).((((.....((((((..(((....)))..)))))).(((.....)))))))....))))).).)))))).))). ( -42.40, z-score = -3.01, R) >droSec1.super_17 1039566 94 + 1527944 AUCGUUCUGUUGGCCACGCA---GAUCU-----------AUUCCGAG-CACAAUGGACCCACAUUGUUCGACCGAUUAACGGCCAAAGACGCGUGCGACGGUAGUCGAC .(((..((((((..(((((.---(.(((-----------....((((-((..(((......))))))))).(((.....)))....)))))))))))))))....))). ( -26.50, z-score = -0.25, R) >droSim1.chrX 3324297 94 - 17042790 AUCGUUCUGUUGGCCACGCA---GAUCU-----------AUUCCGAG-CACAAUGGACCCACAUUGUUCGACCGAUUAACGGCCAAAGACGCGUGCGACGGCAGUCGAC .(((..((((((.((((((.---(.(((-----------....((((-((..(((......))))))))).(((.....)))....))))))))).).)))))).))). ( -29.00, z-score = -1.02, R) >dp4.chrXL_group1e 1492369 100 + 12523060 AUCGUUUCGCUGGCCACGCACACUCGUUGGCCCC-----AUAACGCA-UACAAUGCCCCCGCAUUGUGCUUCUGAUUAACGGGU---CGCGCGUGCAGAAGGAGCUGCC ...(((((.(((..(((((.(((((((((((...-----.....)).-((((((((....))))))))........))))))))---.).))))))))..))))).... ( -32.90, z-score = -0.33, R) >droPer1.super_18 334518 100 + 1952607 AUCGUUUCGCUGGCCACGCACACUCGUUGGCCCC-----AUAACGCA-CACAAUGCCCCCGCAUUGUGCUUCUGAUUAACGGGU---CGCGCGUGCAGAAGGAGCUGCC ...(((((.(((..(((((.(((((((((((...-----.....)).-((((((((....))))))))........))))))))---.).))))))))..))))).... ( -34.60, z-score = -0.76, R) >droAna3.scaffold_13248 1566666 88 - 4840945 AUCGUUCCGUUGGCCACGCAUCGGAAUUAA--------GAUUUUGAGGCACAAUGGAGGUCUAUUGUGGGAGCGAUUAACG----------CGUGCCACAGGGCUC--- .....((((((((((.....(((((((...--------.)))))))))).)))))))(((((..(((((.((((.....))----------).).)))))))))).--- ( -30.80, z-score = -2.13, R) >consensus AUCGUUCUGUUGGCCACGCAGAUCCAUUGG_________AUUCCGAA_CACAAUGGACCCACAUUGUUCGACCGAUUAACGGCCAAACACGCGUGCGACGGCAGUCGAC ........((((..(((((..............................((((((......))))))....(((.....)))........))))))))).......... (-15.50 = -15.12 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:08 2011