
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,181,683 – 17,181,817 |
| Length | 134 |
| Max. P | 0.896984 |

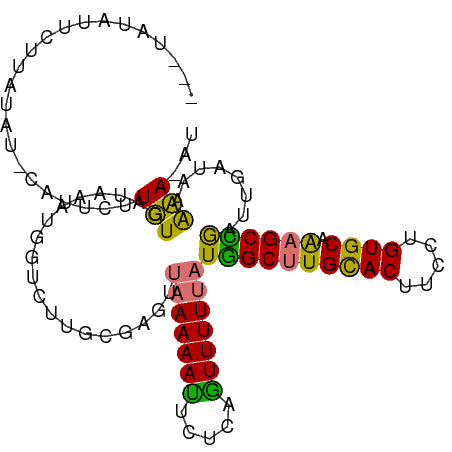
| Location | 17,181,683 – 17,181,786 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.43177 |
| G+C content | 0.30803 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -13.51 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17181683 103 + 22422827 UAUUAUAUUCUUAUAU-UCUUCUUGUGAUAAAUGGUCUUGCGAGCUAAAAAUUCCCGGUUUUUAUGGCUUGCACUUCUUGUGCAAAGCCGAUUGAUAAACAUAU (((((...((......-....((((((((.....)))..))))).(((((((.....))))))).(((((((((.....)))).))))))).)))))....... ( -19.80, z-score = -1.21, R) >droSec1.super_17 1028176 102 + 1527944 UAUUAUAUUCUUAUAUAUAUUCUUGUGAUAAAUGGUCUUAAGAGUUAAAAAUUCU--AUUUUUAUGGCGUGUACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAU ....((((.(((((....(((((((.(((.....))).)))))))..........--........(((.(((((.....)))))..))).....))))).)))) ( -17.00, z-score = -0.37, R) >droYak2.chrX 15796530 88 + 21770863 ---UACAAUUAGCUUU-CCUUCUUUUGAUAAA-----------GUUAAAAACUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAGAGCCAAUUGAUAAAAA-AU ---..(((((.(((((-...((....)).)))-----------))(((((((.....))))))).(((((((((.....))))).))))))))).......-.. ( -21.10, z-score = -3.12, R) >droEre2.scaffold_4690 7522301 94 + 18748788 --------UAUAAUUU-CAUUCUUUUGAUAAAUGCUCUUGCGAGUCAAAAAUUCUCAGUUUUGAUAGCUUGCACUUCCUGUGCAGAGCUGAUUGAUAAAAA-AU --------........-.....(((((((...(((....))).)))))))....((((((.....(((((((((.....))))).))))))))))......-.. ( -20.00, z-score = -1.25, R) >consensus ___UAUAUUCUUAUAU_CAUUCUUGUGAUAAAUGGUCUUGCGAGUUAAAAAUUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCCGAUUGAUAAAAA_AU .............................................(((((((.....)))))))((((((((((.....)))).)))))).............. (-13.51 = -12.82 + -0.69)


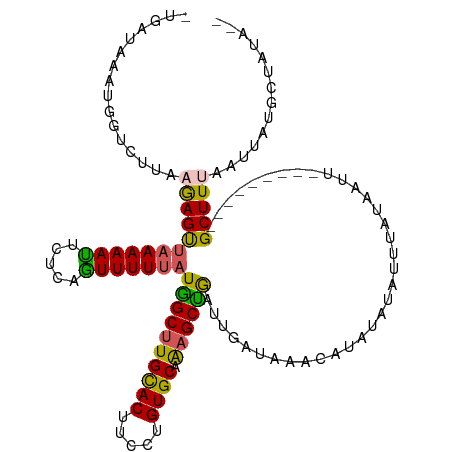
| Location | 17,181,707 – 17,181,817 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.40931 |
| G+C content | 0.30163 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -15.36 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665697 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 17181707 110 + 22422827 -UGAUAAAUGGUCUUGCGAGCUAAAAAUUCCCGGUUUUUAUGGCUUGCACUUCUUGUGCAAAGCCGAUUGAUAAACAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUAUAUA -......(((((.....(((((((((((.....)))))))((((((((((.....)))).))))))..........................---------)))).......)))))... ( -21.10, z-score = -0.64, R) >droSim1.chrX 13355548 109 + 17042790 ACCAUUUAUCGUCUAAAGAGUUAAAAAUUCUCUGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUAUA-- .((((...........((((.........))))......)))).((((((.....))))))((((((((((..((((((((....))))..)---------)))))))))).)))...-- ( -21.53, z-score = -1.37, R) >droSec1.super_17 1028201 106 + 1527944 -UGAUAAAUGGUCUUAAGAGUUAAAAAUUCU--AUUUUUAUGGCGUGUACUUCCUGUGCAAAGCUGAUUGAUAAGCAUAUAUAUUUAUAAUU---------GCUUUAAUUAUGCUAUA-- -.........(((.(((((((..........--))))))).))).(((((.....))))).((((((((((..((((((((....))))..)---------)))))))))).)))...-- ( -19.80, z-score = -0.64, R) >droYak2.chrX 15796551 105 + 21770863 ------------UGAUAAAGUUAAAAACUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAGAGCCAAUUGAUAAA-AAAUAUCUUUACAAUUGCAAGAACUGCUUUACUUAUGCUAUA-- ------------......(((..........((((((((.((((((((((.....))))).)))))...((((..-...))))...........))))))))..........)))...-- ( -23.35, z-score = -2.21, R) >droEre2.scaffold_4690 7522317 116 + 18748788 -UGAUAAAUGCUCUUGCGAGUCAAAAAUUCUCAGUUUUGAUAGCUUGCACUUCCUGUGCAGAGCUGAUUGAUAAA-AAAUAUCGUUAUAAUUCUAAGAAGUGCUUUGCUUAUGCUAUA-- -........((....(((((.((....((((.((..(((.((((((((((.....))))).)))))...((((..-...))))....)))..)).)))).)).)))))....))....-- ( -26.30, z-score = -1.11, R) >consensus _UGAUAAAUGGUCUUAAGAGUUAAAAAUUCUCAGUUUUUAUGGCUUGCACUUCCUGUGCAAAGCUGAUUGAUAAACAUAUAUAUUUAUAAUU_________GCUUUAAUUAUGCUAUA__ ................((((((((((((.....)))))))((((((((((.....)))).))))))...................................))))).............. (-15.36 = -14.76 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:07 2011