
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,133,917 – 17,134,096 |
| Length | 179 |
| Max. P | 0.811920 |

| Location | 17,133,917 – 17,134,023 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.44530 |
| G+C content | 0.50822 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -14.04 |
| Energy contribution | -16.31 |
| Covariance contribution | 2.27 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17133917 106 + 22422827 -----GAUCUAUUAGCGCUGCCUGCGCAU-------CGGGCCAUUUGACUUCAUUUGAUUUCGAGAGCAGACGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGAC -----.........((((.....)))).(-------(((((..(((((..((....))..))))).))....((((((((.....((((....))))..))))))))......)))). ( -34.70, z-score = -2.37, R) >droEre2.scaffold_4690 7475303 113 + 18748788 -----GACCCAUUAGCGCUGCCUGCGCACUUUAGUGCAGGCCAUUUGAAUUCACUUGAUUUCGAGAGCAGAUGCCUGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCAGUCAACCGAC -----.....(((((.((.((((((((......))))))))..((((((.((....)).)))))).......)))))))(((....)))((((((((......))))))))....... ( -42.80, z-score = -4.16, R) >droYak2.chrX 15742135 112 + 21770863 UGGUCGAUCCAUUAGCGCUGCUUGCGCUGC------CGGGCCAUUUGACUUCAUUUGAUUUCGAGAGCAAAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCAGCCGAC ((((..(((...((((((.....)))))).------..(((.(((((.(((...(((....)))))))))))))).))))))).(((((((.(((((......))))).)).))))). ( -38.50, z-score = -1.52, R) >droSec1.super_17 974896 106 + 1527944 -----GAUCUAUUAGCGCUGCCUGCGCAC-------CGGGCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUUCACCGAC -----.........((((.....))))..-------(((((..(((((..((....))..))))).)).(((((((((((.....((((....))))..)))))))))..)).))).. ( -34.00, z-score = -2.30, R) >droSim1.chrX 13306476 106 + 17042790 -----GAUCUAUUAGCGCUGCCUGCGCAC-------CGGGCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGAC -----.........((((.....))))..-------(((((..(((((..((....))..))))).)).(((((((((((.....((((....))))..))))))))).))..))).. ( -34.20, z-score = -2.32, R) >droPer1.super_18 263650 84 + 1952607 -----GAUCCAUUAGCGUUGUAU-UGCUG-------CGAGCCAUUC-----CAUUUGAUUUCGAGAGCAAAUGCCAAUCGGCAGCCC-CGAUCCCCAUUCAAA--------------- -----((((.....((......(-((((.-------((((.((...-----....))..))))..))))).((((....))))))..-.))))..........--------------- ( -16.60, z-score = -0.35, R) >dp4.chrXL_group1e 1424160 84 + 12523060 -----GAUCCAUUAGCGUUGCAU-UGCUG-------CGAGCCAUUC-----CAUUUGAUUUCGAGAGCAAAUGCCAAUCGGCAGCCC-CGAUCCCCAUUCAAA--------------- -----((((.....((......(-((((.-------((((.((...-----....))..))))..))))).((((....))))))..-.))))..........--------------- ( -16.60, z-score = 0.06, R) >consensus _____GAUCCAUUAGCGCUGCCUGCGCAC_______CGGGCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGAC .....((((.....((((.....))))............((((.(((......((((....)))).((....)).......))).))))))))((((......))))........... (-14.04 = -16.31 + 2.27)
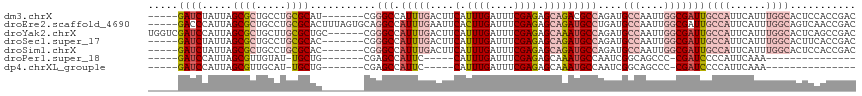
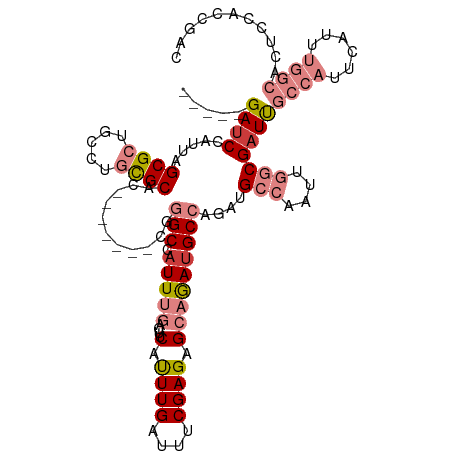

| Location | 17,133,944 – 17,134,057 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Shannon entropy | 0.30183 |
| G+C content | 0.52812 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -21.18 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17133944 113 + 22422827 GCCAUUUGACUUCAUUUGAUUUCGAGAGCAGACGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCCCCGCAGCCCCGUCCACAUCCU------ ((((..(((.....((((....)))).((((.((((((.......)))))).))))...)))..))))........(((..(((.............)))..)))........------ ( -26.92, z-score = -1.52, R) >droEre2.scaffold_4690 7475337 95 + 18748788 GCCAUUUGAAUUCACUUGAUUUCGAGAGCAGAUGCCUGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCAGUCAACCGACACUGCCACCUCAUCCU------------------------ .......(((.((....)).)))(((.((((.((.(....(((....)))((((((((......))))))))....).))))))...))).....------------------------ ( -28.40, z-score = -2.80, R) >droYak2.chrX 15742168 113 + 21770863 GCCAUUUGACUUCAUUUGAUUUCGAGAGCAAAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCAGCCGACACUGCCACUGCCACCCCAUCCUCAUCCUCAUCCU------ ((..(((((..((....))..))))).))...((.(((.((.((.(((((((.(((((......))))).)).)))))...)).))))).)).....................------ ( -24.70, z-score = -1.06, R) >droSec1.super_17 974923 113 + 1527944 GCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUUCACCGACACGGUCACCUCCUCCGCAGCCCCGUCCUCAUCCU------ ((...((((..((....))..))))(((.((.(((((((((.....((((....))))..)))))))))....((((...))))...)).))).)).................------ ( -26.70, z-score = -0.84, R) >droSim1.chrX 13306503 119 + 17042790 GCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCUCCGCAGCCCCACCUCCACCCUCAUCCU (((.(((((..((....))..))))).(.((.(((((((((.....((((....))))..))))))))))).)........)))................................... ( -26.30, z-score = -1.40, R) >consensus GCCAUUUGACUUCAUUUGAUUUCGAGAGCAGAUGCCAGAUGCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCUCCGCAGCCCCAUCCUCAUCCU______ ((((..(((.....((((....)))).((((.((((((.......)))))).))))...)))..))))................................................... (-21.18 = -20.90 + -0.28)

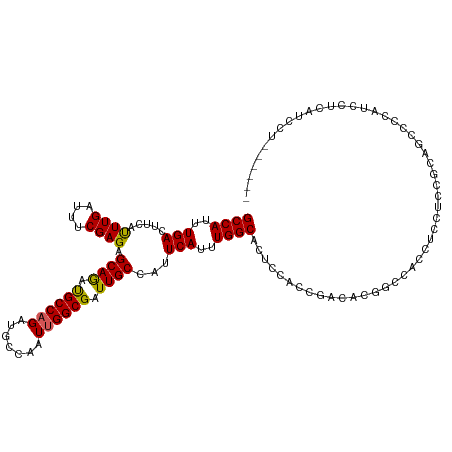
| Location | 17,133,984 – 17,134,096 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Shannon entropy | 0.38352 |
| G+C content | 0.59181 |
| Mean single sequence MFE | -30.79 |
| Consensus MFE | -18.89 |
| Energy contribution | -20.06 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17133984 112 + 22422827 GCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCCCCGCAGCCCCGUC------CACAUCCUCGCCACAGUUCAAUGGCAGUCAUCCACAAUGGAGCCACA ......((((((((((((((.(((((((........(((..(((.............)))..)))------.........)))).)))..)))))))))).(((.....))))))).. ( -32.75, z-score = -2.73, R) >droEre2.scaffold_4690 7475377 90 + 18748788 GCCAAUUGGCGAUUGCCAUUCAUUUGGCAGUCAACCGACACUGCCACCUC------------------------AUCCUCGCCAGAGUUCGAUGGCAG----CCACAAUGGAGCCGCA .(((..((((((((((((......)))))))).........(((((..((------------------------...(((....)))...))))))))----)))...)))....... ( -31.10, z-score = -2.33, R) >droYak2.chrX 15742208 97 + 21770863 GCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCAGCCGACACUGCCACUGCCACCCCAUCCUCAUC------CUCAUCCUGGCCAGAGUUCGAUGGCAG----CCACA----------- ((...(((((((.(((((......))))).)).)))))....))..((((((.(.....(((..(------(.......))...)))...).))))))----.....----------- ( -27.20, z-score = -1.27, R) >droSec1.super_17 974963 112 + 1527944 GCCAAUUGGCGAUUGCCAUUCAUUUGGCACUUCACCGACACGGUCACCUCCUCCGCAGCCCCGUC------CUCAUCCUCGCCAGAGUUCAGUGGCAGCCAUCCACAAUGGAGCCGCA .....(((((((.(((((......))))).......(((..(((..(.......)..)))..)))------.......)))))))......(((((..((((.....)))).))))). ( -32.00, z-score = -1.23, R) >droSim1.chrX 13306543 118 + 17042790 GCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCUCCGCAGCCCCACCUCCACCCUCAUCCUCGCCAGAGUUCAGUGGCAGCCAUCCACAAUGGAGCCGCA (((..((((.((.(((((......))))).))..))))...)))..........((.((.(((..............(((....)))....((((.......))))..))).)).)). ( -30.90, z-score = -1.05, R) >consensus GCCAAUUGGCGAUUGCCAUUCAUUUGGCACUCCACCGACACGGCCACCUCCUCCGCAGCCCCAUC______CUCAUCCUCGCCAGAGUUCAAUGGCAG_CAUCCACAAUGGAGCCGCA (((....)))((.(((((......))))).)).........(((....................................((((........)))).....(((.....))))))... (-18.89 = -20.06 + 1.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:00 2011