
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,129,805 – 17,129,885 |
| Length | 80 |
| Max. P | 0.861176 |

| Location | 17,129,805 – 17,129,885 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.53711 |
| G+C content | 0.34660 |
| Mean single sequence MFE | -14.77 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
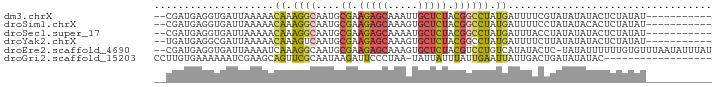
>dm3.chrX 17129805 80 + 22422827 -----------AUAUAGAGUAUAUAUACGAAAAUCAUAGGCCGUAGAGCAAUUUGCUCUUCGCAUUGCCUUUGUUUUUAAUCACCUCAUCG-- -----------.....(((.........(((((.((.((((((.(((((.....))))).))....)))).)))))))......)))....-- ( -16.96, z-score = -2.31, R) >droSim1.chrX 13302347 80 + 17042790 -----------AUAUAGAGUGUAUAUAGGAAAAUCAUAGGCCGUAGAGCACUUUGCUCUUCGCAUUGCCUUUGUUUUUAAUCACCUCAUCG-- -----------.....(((.((......(((((.((.((((((.(((((.....))))).))....)))).)))))))....)))))....-- ( -20.00, z-score = -2.59, R) >droSec1.super_17 965768 80 + 1527944 -----------AUAUAGAGUAUAUAUAGGUAAAUCAUAGGCCGUAGAGCAUUUUGCUCUUCGCAUUGCCUUUGUUUUUAAUCACCUCAUCG-- -----------(((((.....)))))(((((((.((.((((((.(((((.....))))).))....)))).)).))).....)))).....-- ( -17.00, z-score = -2.04, R) >droYak2.chrX 15737809 80 + 21770863 -----------AUAUAGAGUAUAUAUAAGAAAAUCAUAGGCCGUAGAGCACUUUGCUCUUCGCAUUGACUUUGUUUUUAAUCGCCUCAUCA-- -----------.....(((.......((.(((.(((...((...(((((.....)))))..))..))).))).)).........)))....-- ( -14.39, z-score = -0.80, R) >droEre2.scaffold_4690 7471188 90 + 18748788 AUAAAUAUUAAACACAAAAAAUAUA-GAGUAUAUGACAGGACGUAGAGCACUUUGCUCUUCGCAUUGCCUUUGAUUUUAAUCACCUCAUCG-- .......................((-((((....(.(((..((.(((((.....))))).))..))).)....))))))............-- ( -13.80, z-score = -1.10, R) >droGri2.scaffold_15203 5523367 74 - 11997470 ------------------GUAUAUAUCAGUCAAUAAUUCAAUAAAUAAUA-UUAGGGAAUCUUAUUGCGAACUGCUUCGAUUUUUUCACAAGG ------------------........(((((((((((((..(((......-)))..))))..)))))...))))................... ( -6.50, z-score = 0.31, R) >consensus ___________AUAUAGAGUAUAUAUAAGAAAAUCAUAGGCCGUAGAGCACUUUGCUCUUCGCAUUGCCUUUGUUUUUAAUCACCUCAUCG__ ..................................((.((((((.(((((.....))))).))....)))).)).................... ( -6.96 = -7.60 + 0.64)



| Location | 17,129,805 – 17,129,885 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.53711 |
| G+C content | 0.34660 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -8.27 |
| Energy contribution | -8.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17129805 80 - 22422827 --CGAUGAGGUGAUUAAAAACAAAGGCAAUGCGAAGAGCAAAUUGCUCUACGGCCUAUGAUUUUCGUAUAUAUACUCUAUAU----------- --....(((...((.((((.((.((((....((.(((((.....))))).)))))).)).)))).)).......))).....----------- ( -17.10, z-score = -1.57, R) >droSim1.chrX 13302347 80 - 17042790 --CGAUGAGGUGAUUAAAAACAAAGGCAAUGCGAAGAGCAAAGUGCUCUACGGCCUAUGAUUUUCCUAUAUACACUCUAUAU----------- --....((((((((.((((.((.((((....((.(((((.....))))).)))))).)).))))...)).))).))).....----------- ( -18.30, z-score = -1.95, R) >droSec1.super_17 965768 80 - 1527944 --CGAUGAGGUGAUUAAAAACAAAGGCAAUGCGAAGAGCAAAAUGCUCUACGGCCUAUGAUUUACCUAUAUAUACUCUAUAU----------- --.....((((((.......((.((((....((.(((((.....))))).)))))).))..))))))...............----------- ( -18.70, z-score = -2.85, R) >droYak2.chrX 15737809 80 - 21770863 --UGAUGAGGCGAUUAAAAACAAAGUCAAUGCGAAGAGCAAAGUGCUCUACGGCCUAUGAUUUUCUUAUAUAUACUCUAUAU----------- --.....((((((((........))))....((.(((((.....))))).))))))..........................----------- ( -16.80, z-score = -1.56, R) >droEre2.scaffold_4690 7471188 90 - 18748788 --CGAUGAGGUGAUUAAAAUCAAAGGCAAUGCGAAGAGCAAAGUGCUCUACGUCCUGUCAUAUACUC-UAUAUUUUUUGUGUUUAAUAUUUAU --....(((.((((....))))..((((..(((.(((((.....))))).)))..)))).....)))-......................... ( -19.40, z-score = -1.51, R) >droGri2.scaffold_15203 5523367 74 + 11997470 CCUUGUGAAAAAAUCGAAGCAGUUCGCAAUAAGAUUCCCUAA-UAUUAUUUAUUGAAUUAUUGACUGAUAUAUAC------------------ ..(((((((.............)))))))...(((((..(((-......)))..)))))................------------------ ( -6.22, z-score = 0.44, R) >consensus __CGAUGAGGUGAUUAAAAACAAAGGCAAUGCGAAGAGCAAAGUGCUCUACGGCCUAUGAUUUACUUAUAUAUACUCUAUAU___________ ....................((.((((....((.(((((.....))))).)))))).)).................................. ( -8.27 = -8.33 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:58 2011