| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,115,219 – 17,115,310 |
| Length | 91 |
| Max. P | 0.997346 |

| Location | 17,115,219 – 17,115,310 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.31 |
| Shannon entropy | 0.56704 |
| G+C content | 0.56515 |
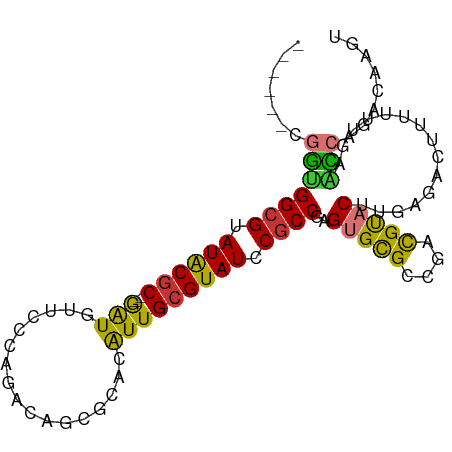
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -18.10 |
| Energy contribution | -16.89 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
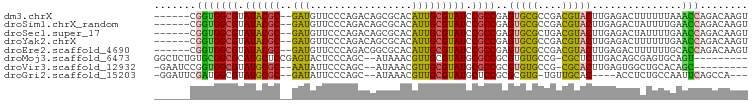
>dm3.chrX 17115219 91 + 22422827 ------CGGUGGCGUAUACGC--GAUGUUCCCAGACAGCGCACAUUGCGUAUCCGCCGAGUGCGCCGACGUACUUGAGACUUUUUUAAACCAGACAAGU ------.(((((((.((((((--(((((.(.(.....).).))))))))))).))))(((((((....))))))).............)))........ ( -33.40, z-score = -3.23, R) >droSim1.chrX_random 4399817 91 + 5698898 ------CGGUGGCGUAUACGC--GAUGUUCCCAGACAGCGCACAUUGCGUAUCCGCCGAGUGCGCCGACGUACUUGAGACUAUUUUGAACCAGACAAGU ------.(((((((.((((((--(((((.(.(.....).).))))))))))).))))(((((((....))))))).............)))........ ( -33.40, z-score = -2.97, R) >droSec1.super_17 951574 91 + 1527944 ------CGGUGGCGUAUACGC--GAUGUUCCCAGACAGCGCACAUUGCGUAUCCGCCGAGUGCGCUGACGUACUUGAGACUAUUUUGAACCAGACAAGU ------.(((((((.((((((--(((((.(.(.....).).))))))))))).))))(((((((....))))))).............)))........ ( -33.40, z-score = -2.86, R) >droYak2.chrX 15721862 91 + 21770863 ------CGGUGGCGUAUACGC--GAUGUUCCCAGACAGCGCACAUUGCGUAUCCGCCGAGUGCGCCGACGUACUUGAGACUUUUUUGAACCAGACAAGU ------.(((((((.((((((--(((((.(.(.....).).))))))))))).))))(((((((....))))))).............)))........ ( -33.40, z-score = -2.95, R) >droEre2.scaffold_4690 7456855 91 + 18748788 ------CGGUGGCGUAUACGC--GAUGUUCCCAGACGGCGCACAUUGCGUAUCCGCCGAGUGCGCCGACGUACUUGAGACUUUUUUGCACCAGACAAGU ------.(((((((.((((((--(((((.(((....)).).))))))))))).)))((((((((....))))))))...........))))........ ( -37.90, z-score = -3.45, R) >droMoj3.scaffold_6473 5586369 87 + 16943266 GGCUCUGUGCGGCGCAUGCUCCGAGUACUCCCAGC--AUAAACGUUGCGUAUGCGCCGCGUGUGCCG-CGCUCUUGACAGCGAGUGCAGU--------- (((.(...(((((((((((...((....))...((--(.......))))))))))))))..).)))(-(((((........))))))...--------- ( -40.30, z-score = -1.87, R) >droVir3.scaffold_12932 2024039 84 + 2102469 -GAAUCCGGUGGCGUAUGCGC--AAUAUUCCCAGC--AUAAACGUUGCGUAUGCGCCGCGUGUGCCG-CGCACUUGAGUGGCUGCACAGC--------- -.......(((((((((((((--(((.........--......))))))))))))))))((((((((-(........))))).))))...--------- ( -39.96, z-score = -2.70, R) >droGri2.scaffold_15203 5504260 86 - 11997470 -GGAUUCGAUGGCGUAUGCGC--GAUAUUCCCAGC--AUAAACGUUGCGUAUGCUCCGCGCGUG-UGUUGCAC----ACCUCUGCCAAUUCAGCCA--- -((......((((((((((((--(((.........--......))))))))))).......(((-((...)))----))....))))......)).--- ( -25.46, z-score = 0.02, R) >consensus ______CGGUGGCGUAUACGC__GAUGUUCCCAGACAGCGCACAUUGCGUAUCCGCCGAGUGCGCCGACGUACUUGAGACUUUUUUGAACCAGACAAGU .......(((((((.((((((....(((.........)))......)))))).))))..(((((....)))))...............)))........ (-18.10 = -16.89 + -1.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:54 2011