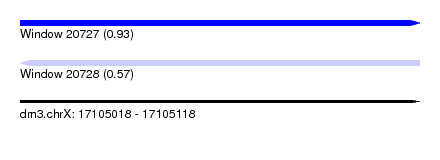
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,105,018 – 17,105,118 |
| Length | 100 |
| Max. P | 0.933047 |

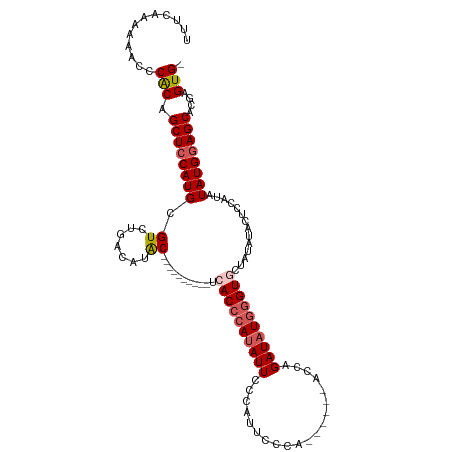
| Location | 17,105,018 – 17,105,118 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.66 |
| Shannon entropy | 0.27771 |
| G+C content | 0.44864 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -16.23 |
| Energy contribution | -17.27 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17105018 100 + 22422827 UUUAAAAAAACCCACAGCUCCAUGCGUCUGACAUAC----------UCACCAAUAUUCCCAUUCCCA------ACAAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCACGAGUG- ................((((((((....(((.....----------)))...((((.(((((((...------....)).)))))....))))........)))))))).......- ( -18.00, z-score = -1.17, R) >droSim1.chrX 13279754 100 + 17042790 UUUCAAAAAACCCACAGCUCCAUGCGUCUGACAUGC----------UCACCCAUAUUCCCAUUCCCA------ACUAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCGCGAGUG- ............(((.(((((((((((.....))))----------.((((((((((..........------....))))))))))...............)))))))....)))- ( -26.14, z-score = -3.10, R) >droSec1.super_17 941250 100 + 1527944 UUUAAAAAAACCCACAGCUCCAUGCGUCUGACAUGC----------UCACCCAAAUUCCCAUUCCCA------ACUAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCGCGAGUG- ............(((.(((((((((((.....))))----------.((((((.(((..........------....))).))))))...............)))))))....)))- ( -22.04, z-score = -1.94, R) >droYak2.chrX 15711445 116 + 21770863 UUCCACAAAAUUCGCAGCUCCAUGCGUCUGACACACGACGACACACUCACCCAUAUUCCAAUUUCCAUUCCCAAACAGAUAUGAGUACUAUAUGCUCCAUAUAUGGAGCACGAGUG- .............(((......)))(((........)))....(((((((.((((((....................)))))).))......(((((((....))))))).)))))- ( -24.95, z-score = -3.12, R) >droEre2.scaffold_4690 7446655 114 + 18748788 UUCCAAAAAAUCCACAGCUCCAUGCGUCUGACACACGAC---ACACUCACCCAUAUUCCCAUUCCCAUUCCCAACCAGAUAUGGGUACUAUCUACUCCAUAUAUGAAGCACGAGUGG ...........((((.(((.((((.(((........)))---......(((((((((....................)))))))))...............)))).)))....)))) ( -20.25, z-score = -1.79, R) >consensus UUUCAAAAAACCCACAGCUCCAUGCGUCUGACAUAC__________UCACCCAUAUUCCCAUUCCCA______ACCAGAUAUGGGUGCUAUAUACUCCAUAUAUGGAGCACGAGUG_ ............(((.((((((((.(.....)...............((((((((((....................))))))))))..............))))))))....))). (-16.23 = -17.27 + 1.04)

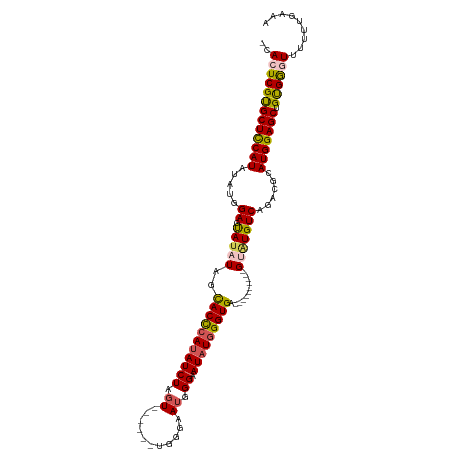
| Location | 17,105,018 – 17,105,118 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.66 |
| Shannon entropy | 0.27771 |
| G+C content | 0.44864 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17105018 100 - 22422827 -CACUCGUGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUUGU------UGGGAAUGGGAAUAUUGGUGA----------GUAUGUCAGACGCAUGGAGCUGUGGGUUUUUUUAAA -.((((((((((((....))))))))(((((.(((((.(((...------.))).)))))....((.(((.----------((.......)).))).)))))))))))......... ( -29.70, z-score = -1.64, R) >droSim1.chrX 13279754 100 - 17042790 -CACUCGCGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGU------UGGGAAUGGGAAUAUGGGUGA----------GCAUGUCAGACGCAUGGAGCUGUGGGUUUUUUGAAA -.(((((((((((((...............((((((((((((..------......)))..))))))))).----------((.........))))))))).))))))......... ( -32.00, z-score = -1.80, R) >droSec1.super_17 941250 100 - 1527944 -CACUCGCGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGU------UGGGAAUGGGAAUUUGGGUGA----------GCAUGUCAGACGCAUGGAGCUGUGGGUUUUUUUAAA -.(((((((((((((.((((.....))))((.(((((.(((...------.))).))))).....(..(((----------.....)))..)))))))))).))))))......... ( -30.10, z-score = -1.45, R) >droYak2.chrX 15711445 116 - 21770863 -CACUCGUGCUCCAUAUAUGGAGCAUAUAGUACUCAUAUCUGUUUGGGAAUGGAAAUUGGAAUAUGGGUGAGUGUGUCGUCGUGUGUCAGACGCAUGGAGCUGCGAAUUUUGUGGAA -(((((((((((((....)))))))).....((((((((((((((........))).))).))))))))))))).....(((((((.....)))))))..(..(((...)))..).. ( -34.50, z-score = -1.31, R) >droEre2.scaffold_4690 7446655 114 - 18748788 CCACUCGUGCUUCAUAUAUGGAGUAGAUAGUACCCAUAUCUGGUUGGGAAUGGGAAUGGGAAUAUGGGUGAGUGU---GUCGUGUGUCAGACGCAUGGAGCUGUGGAUUUUUUGGAA ((((....(((((.................(((((((((((.(((.........))).)).))))))))).((((---(((........)))))))))))).))))........... ( -34.90, z-score = -1.60, R) >consensus _CACUCGUGCUCCAUAUAUGGAGUAUAUAGCACCCAUAUCUAGU______UGGGAAUGGGAAUAUGGGUGA__________GUAUGUCAGACGCAUGGAGCUGUGGGUUUUUUGAAA .(((((((((((((....))))))))......(((((.((((........)))).))))).....))))).....................((((......))))............ (-23.68 = -23.92 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:53 2011