
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,059,515 – 17,059,644 |
| Length | 129 |
| Max. P | 0.883868 |

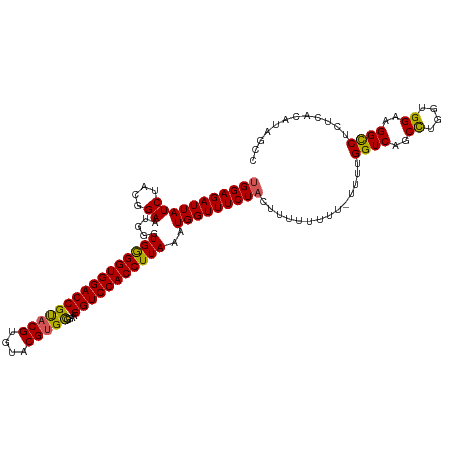
| Location | 17,059,515 – 17,059,633 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.81 |
| Shannon entropy | 0.12512 |
| G+C content | 0.49496 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -35.24 |
| Energy contribution | -34.76 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17059515 118 - 22422827 UGGAGAUUAUCUACGGAAUGGGUGAGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU-UUUUGGUCAGCCUGGUGGAAGGCCUCUCACAUAGCC (((((((((((....)).....((((((((((((((((....)))))....)))))))))))..))))))))).........-....((((..((....))..))))............ ( -40.10, z-score = -1.87, R) >droEre2.scaffold_4690 7405872 118 - 18748788 UGGAGAUUAUCUAC-GAAUGGGUGGGGUGGACCGUACGUGUGCGUGUGGUAGGUCCACCUUAAAUGGUUUCUACUUUUUUUUGUAUUGGUCAGCUUGGUGGAAGGCCCGUCACAUAGCC ((((.....)))).-((.(((((((((((((((.((((....)))).....))))))))))......((((((((.....(((.......)))...)))))))))))))))........ ( -36.00, z-score = -0.47, R) >droYak2.chrX 15669053 115 - 21770863 UGGAGAUUAUCUAC-GAAUGGGUGGGGUGGACCGCACGUGUGCGUGUGGUAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU---UUGGUCAGCCUGGCGGAAGGCCUCUCACAUAGCC ((((((((((.((.-....(((((((.(..((((((((....)))))))).).))))))))).)))))))))).........---..((((..((....))..))))............ ( -38.90, z-score = -1.07, R) >droSec1.super_17 901177 116 - 1527944 CGGAGAUUAUCUACGGAAUGGGUGGGGUGGACCGU-CGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUAC-UUUUUUU-UUUUGGUCAGCCUGGUGGAAGGUCUCUCACAUAGCC .((((((..((((((((((.(.(((((((((((.(-(((......))))..)))))))))))..).)))))...-.......-....((....))..)))))..))))))......... ( -40.60, z-score = -2.08, R) >droSim1.chrX 13241802 117 - 17042790 UGGAGAUUAUCUACGGAAUGGAUGGGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUAC-UUUUUUU-UUUUGGUCAGCCUGGUGGAAGGCCUCUCACAUAGCC (((((((((((....)).....((((((((((((((((....)))))....)))))))))))..))))))))).-.......-....((((..((....))..))))............ ( -39.40, z-score = -1.65, R) >consensus UGGAGAUUAUCUACGGAAUGGGUGGGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU_UUUUGGUCAGCCUGGUGGAAGGCCUCUCACAUAGCC (((((((((((....)).....((((((((((((((((....)))))....)))))))))))..)))))))))..............((((..((....))..))))............ (-35.24 = -34.76 + -0.48)

| Location | 17,059,526 – 17,059,644 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Shannon entropy | 0.12512 |
| G+C content | 0.49665 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17059526 118 + 22422827 AGGCCUUCCACCAGGCUGACCAAAA-AAAAAAAAGUAGAAACCAUUUAAGGUGGACCUUCCGCACGUACACGUACGGUCCACCUCACCCAUUCCGUAGAUAAUCUCCAUUGGCAUCUGC .(((((......)))))........-........(((((..(((....(((((((((....(.(((....))).))))))))))..........(.((.....)).)..)))..))))) ( -30.00, z-score = -2.19, R) >droEre2.scaffold_4690 7405883 118 + 18748788 GGGCCUUCCACCAAGCUGACCAAUACAAAAAAAAGUAGAAACCAUUUAAGGUGGACCUACCACACGCACACGUACGGUCCACCCCACCCAUUC-GUAGAUAAUCUCCACUGGCAUCUGC ((........))......................(((((..(((.....(((((((((((...........))).))))))))..........-((.((.....)).)))))..))))) ( -24.70, z-score = -0.83, R) >droYak2.chrX 15669064 115 + 21770863 AGGCCUUCCGCCAGGCUGACCAA---AAAAAAAAGUAGAAACCAUUUAAGGUGGACCUACCACACGCACACGUGCGGUCCACCCCACCCAUUC-GUAGAUAAUCUCCAUUGGCAUCUGC .(((((......)))))......---........(((((..(((.....((((((((.....((((....)))).))))))))..........-(.((.....)).)..)))..))))) ( -31.00, z-score = -2.03, R) >droSec1.super_17 901188 116 + 1527944 AGACCUUCCACCAGGCUGACCAAAA-AAAAAAA-GUAGAAACCAUUUAAGGUGGACCUUCCGCACGUACACG-ACGGUCCACCCCACCCAUUCCGUAGAUAAUCUCCGUUGGCAUCUGC ((.(((......)))))........-.......-(((((..(((.....((((((((.((...........)-).))))))))..........((.((.....)).)).)))..))))) ( -25.60, z-score = -1.02, R) >droSim1.chrX 13241813 117 + 17042790 AGGCCUUCCACCAGGCUGACCAAAA-AAAAAAA-GUAGAAACCAUUUAAGGUGGACCUUCCGCACGUACACGUACGGUCCACCCCAUCCAUUCCGUAGAUAAUCUCCAUUGGCAUCUGC .(((((......)))))........-.......-(((((..(((.....((((((((....(.(((....))).)))))))))...........(.((.....)).)..)))..))))) ( -29.00, z-score = -1.92, R) >consensus AGGCCUUCCACCAGGCUGACCAAAA_AAAAAAAAGUAGAAACCAUUUAAGGUGGACCUUCCGCACGUACACGUACGGUCCACCCCACCCAUUCCGUAGAUAAUCUCCAUUGGCAUCUGC .(((((......))))).................(((((..(((.....((((((((......(((....)))..))))))))...........(.((.....)).)..)))..))))) (-25.00 = -25.60 + 0.60)



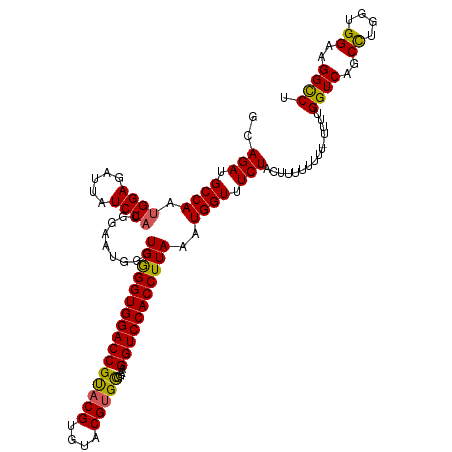
| Location | 17,059,526 – 17,059,644 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.81 |
| Shannon entropy | 0.12512 |
| G+C content | 0.49665 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -37.22 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17059526 118 - 22422827 GCAGAUGCCAAUGGAGAUUAUCUACGGAAUGGGUGAGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU-UUUUGGUCAGCCUGGUGGAAGGCCU ..(((.((((..........((....)).....((((((((((((((((....)))))....)))))))))))..)))).)))..........-....((((..((....))..)))). ( -41.20, z-score = -2.16, R) >droEre2.scaffold_4690 7405883 118 - 18748788 GCAGAUGCCAGUGGAGAUUAUCUAC-GAAUGGGUGGGGUGGACCGUACGUGUGCGUGUGGUAGGUCCACCUUAAAUGGUUUCUACUUUUUUUUGUAUUGGUCAGCUUGGUGGAAGGCCC (((((....((((((((((((((((-......))))((((((((.((((....)))).....))))))))....))))))))))))....)))))...((((..((....))..)))). ( -41.20, z-score = -1.96, R) >droYak2.chrX 15669064 115 - 21770863 GCAGAUGCCAAUGGAGAUUAUCUAC-GAAUGGGUGGGGUGGACCGCACGUGUGCGUGUGGUAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU---UUGGUCAGCCUGGCGGAAGGCCU ((...(((((.((((((((((.((.-....(((((((.(..((((((((....)))))))).).))))))))).)))))))))).........---..((....)))))))....)).. ( -42.10, z-score = -1.86, R) >droSec1.super_17 901188 116 - 1527944 GCAGAUGCCAACGGAGAUUAUCUACGGAAUGGGUGGGGUGGACCGU-CGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUAC-UUUUUUU-UUUUGGUCAGCCUGGUGGAAGGUCU ((....))......((((..((((((((((.(.(((((((((((.(-(((......))))..)))))))))))..).)))))...-.......-....((....))..)))))..)))) ( -36.50, z-score = -0.65, R) >droSim1.chrX 13241813 117 - 17042790 GCAGAUGCCAAUGGAGAUUAUCUACGGAAUGGAUGGGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUAC-UUUUUUU-UUUUGGUCAGCCUGGUGGAAGGCCU ..(((.((((..........((....)).....((((((((((((((((....)))))....)))))))))))..)))).)))..-.......-....((((..((....))..)))). ( -40.50, z-score = -1.96, R) >consensus GCAGAUGCCAAUGGAGAUUAUCUACGGAAUGGGUGGGGUGGACCGUACGUGUACGUGCGGAAGGUCCACCUUAAAUGGUUUCUACUUUUUUUU_UUUUGGUCAGCCUGGUGGAAGGCCU ..(((.((((.((((.....)))).........((((((((((((((((....)))))....)))))))))))..)))).)))...............((((..((....))..)))). (-37.22 = -36.74 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:48 2011