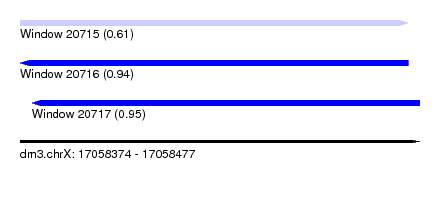
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,058,374 – 17,058,477 |
| Length | 103 |
| Max. P | 0.952518 |

| Location | 17,058,374 – 17,058,474 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 72.38 |
| Shannon entropy | 0.46921 |
| G+C content | 0.30784 |
| Mean single sequence MFE | -18.22 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
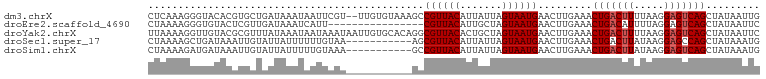
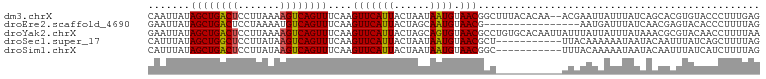
>dm3.chrX 17058374 100 + 22422827 CUCAAAGGGUACACGUGCUGAUAAAUAAUUCGU--UUGUGUAAAGCCGUUACAUUAUUAGUAAUGAACUUGAAACUGACUUUUAAGGAGUCAGCUAUAAUUG .((((.((.((((((.((.((........))))--.))))))...))(((.(((((....))))))))))))..(((((((.....)))))))......... ( -22.70, z-score = -1.81, R) >droEre2.scaffold_4690 7404777 86 + 18748788 CUAAAAGGGUGUACUCGUUGAUAAAUCAUU----------------CGUUACAUUGCUAGUAAUGAACUUGAAACUGACAUUUUAGGAGUCAGCUAUAAUUC ......(((....)))((((((......((----------------((((((.......))))))))(((((((......))))))).))))))........ ( -17.70, z-score = -1.34, R) >droYak2.chrX 15667956 102 + 21770863 UUAAAAGGUUGUACGCGUUUAUAAAUAAUAAAUAAUUGUGCACAGGCGUUACACUGCUAGUAAUGAACUUGAAACUGACUUUUAAGGAGUCAGCUAUAAUUC .........((((((.((((((.....))))))...))))))((((((((((.......))))))..))))...(((((((.....)))))))......... ( -20.30, z-score = -0.85, R) >droSec1.super_17 900053 91 + 1527944 CUAAAAGCUGAUAAAUUGUAUUAUUUUUUGUAA-----------AGCGUUACAUUAUUAGUAAUGAACUUGAAACUGACUUAUAAGGAGCCAGCUAUAAAUG .....(((((.....((((((((((((..((..-----------..((((((.......)))))).))..)))).)))..))))).....)))))....... ( -13.40, z-score = -0.32, R) >droSim1.chrX 13240680 91 + 17042790 CUAAAAGAUGAUAAAUUGUAUUAUUUUUGUAAA-----------GCCGUUACAUUAUUAGUAAUGAACUUGAAACUGACUUAUAAGGAGUCAGCUAUAAAUG ...((((((((((.....)))))))))).....-----------..((((((.......)))))).........(((((((.....)))))))......... ( -17.00, z-score = -2.11, R) >consensus CUAAAAGGUUAUACAUGGUAAUAAAUAAUUAAA___________GCCGUUACAUUAUUAGUAAUGAACUUGAAACUGACUUUUAAGGAGUCAGCUAUAAUUG ..............................................((((((.......)))))).........(((((((.....)))))))......... (-10.20 = -10.60 + 0.40)
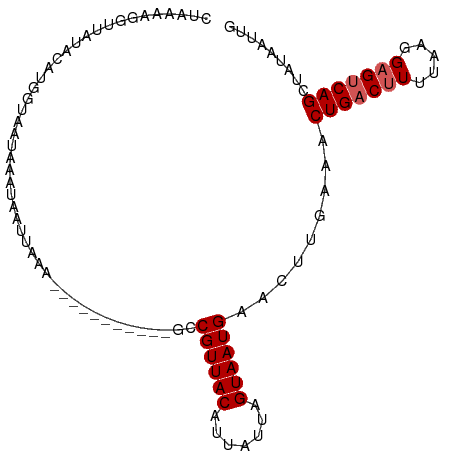
| Location | 17,058,374 – 17,058,474 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Shannon entropy | 0.46921 |
| G+C content | 0.30784 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17058374 100 - 22422827 CAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGGCUUUACACAA--ACGAAUUAUUUAUCAGCACGUGUACCCUUUGAG .......((((((((.......))))))))((((((((((((....))))).)))((...(((((..--...................))))).)).)))). ( -16.80, z-score = -1.45, R) >droEre2.scaffold_4690 7404777 86 - 18748788 GAAUUAUAGCUGACUCCUAAAAUGUCAGUUUCAAGUUCAUUACUAGCAAUGUAACG----------------AAUGAUUUAUCAACGAGUACACCCUUUUAG (((((..(((((((.........)))))))...)))))...........((((...----------------..(((....))).....))))......... ( -13.80, z-score = -1.31, R) >droYak2.chrX 15667956 102 - 21770863 GAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAGCAGUGUAACGCCUGUGCACAAUUAUUUAUUAUUUAUAAACGCGUACAACCUUUUAA (((((..((((((((.......))))))))...))))).......((.(((((.......))))).....(((((.....))))).)).............. ( -18.80, z-score = -1.76, R) >droSec1.super_17 900053 91 - 1527944 CAUUUAUAGCUGGCUCCUUAUAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGCU-----------UUACAAAAAAUAAUACAAUUUAUCAGCUUUUAG .......((((((((.......))))))))....((((((((....))))).)))...-----------................................. ( -11.00, z-score = -0.77, R) >droSim1.chrX 13240680 91 - 17042790 CAUUUAUAGCUGACUCCUUAUAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGGC-----------UUUACAAAAAUAAUACAAUUUAUCAUCUUUUAG .......((((((((.......))))))))....((((((((....))))).)))...-----------................................. ( -11.60, z-score = -1.73, R) >consensus CAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGCC___________AUUAAUAAUUUAUCAACACGUAUAACCUUUUAG .......((((((((.......))))))))....(((((((......)))).)))............................................... (-10.36 = -10.24 + -0.12)
| Location | 17,058,377 – 17,058,477 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.48426 |
| G+C content | 0.30560 |
| Mean single sequence MFE | -14.74 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
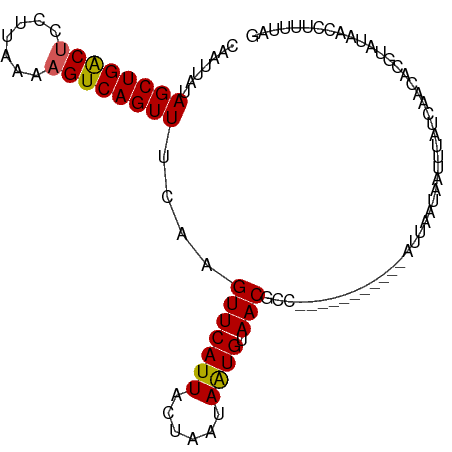
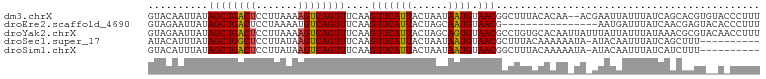
>dm3.chrX 17058377 100 - 22422827 GUACAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGGCUUUACACAA--ACGAAUUAUUUAUCAGCACGUGUACCCUUU (((((.....((((((((.......))))))))....((((((((....))))).)))............--....................)))))..... ( -16.10, z-score = -1.46, R) >droEre2.scaffold_4690 7404780 86 - 18748788 GUAGAAUUAUAGCUGACUCCUAAAAUGUCAGUUUCAAGUUCAUUACUAGCAAUGUAACG----------------AAUGAUUUAUCAACGAGUACACCCUUU ...(((((..(((((((.........)))))))...)))))...........((((...----------------..(((....))).....))))...... ( -14.00, z-score = -1.11, R) >droYak2.chrX 15667959 102 - 21770863 GUAGAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAGCAGUGUAACGCCUGUGCACAAUUAUUUAUUAUUUAUAAACGCGUACAACCUUU ((((((((..((((((((.......))))))))...))))).......((.(((((.......))))).....(((((.....))))).)).)))....... ( -20.10, z-score = -1.89, R) >droSec1.super_17 900056 91 - 1527944 AUACAUUUAUAGCUGGCUCCUUAUAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGCUUUACAAAAAAUA-AUACAAUUUAUCAGCUUU---------- ..........((((((((.......))))))))....((((((((....))))).)))...............-..................---------- ( -11.00, z-score = -0.87, R) >droSim1.chrX 13240683 91 - 17042790 GUACAUUUAUAGCUGACUCCUUAUAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGGCUUUACAAAAAUA-AUACAAUUUAUCAUCUUU---------- (((.......((((((((.......))))))))....((((((((....))))).)))...............-.)))..............---------- ( -12.50, z-score = -1.96, R) >consensus GUACAAUUAUAGCUGACUCCUUAAAAGUCAGUUUCAAGUUCAUUACUAAUAAUGUAACGCCUUUACAAAA_UA_AUAUAAUUUAUCAACGCGU__A_CCUUU ..........((((((((.......))))))))....(((((((......)))).)))............................................ (-10.36 = -10.24 + -0.12)
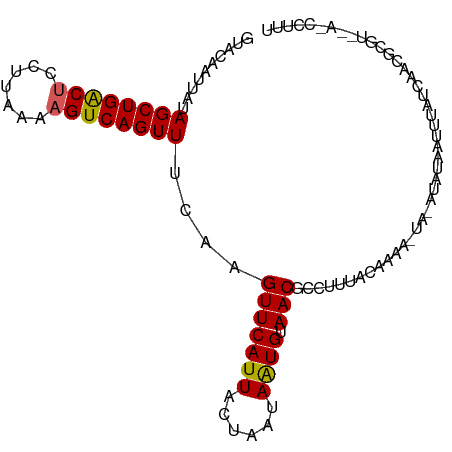
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:44 2011