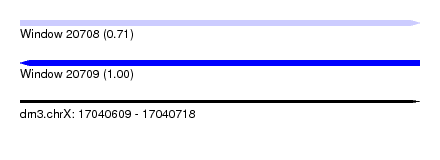
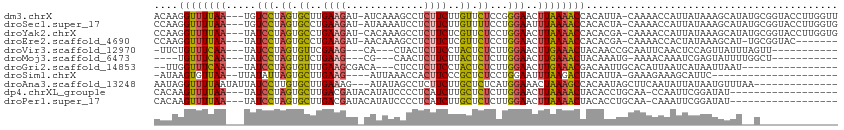
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,040,609 – 17,040,718 |
| Length | 109 |
| Max. P | 0.997672 |

| Location | 17,040,609 – 17,040,718 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 61.91 |
| Shannon entropy | 0.75938 |
| G+C content | 0.37023 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -9.67 |
| Energy contribution | -8.92 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.710006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
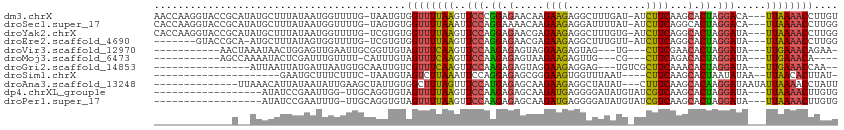
>dm3.chrX 17040609 109 + 22422827 AACCAAGGUACCGCAUAUGCUUUAUAAUGGUUUUG-UAAUGUGGUUUUAAGUUCCCGGAGAACAAGAAGAGGCUUUGAU-AUCUUCAAGCACUAGGACA---UUAAAACCUUGU ...(((((((((((((.(((..............)-)))))))))(((((..(((..(.(.....(((((.........-.)))))...).)..)))..---))))))))))). ( -24.84, z-score = -0.76, R) >droSec1.super_17 881707 109 + 1527944 CACCAAGGUACCGCAUAUGCUUUAUAAUGGUUUUG-UAGUGUGGUUUUAAAUUCCAGGAAAACAAGAAGAGGAUUUUAU-AUCUUCAGGCACUAGGACA---UUAAAACCUUGG ..(((((((...((....))....(((((...((.-((((((.(((((..........))))).....((((((.....-))))))..)))))).))))---)))..))))))) ( -28.00, z-score = -2.16, R) >droYak2.chrX 15650330 109 + 21770863 CACCAAGGUACCGCAUAUGCUUUAUAAUGGUUUUG-UCGUGUGGUUUUAAGUUCCAGGAGAACGAGAAGAGGCUUUGUG-AUCUUCAGGCACUAGGAUA---UUAAAACCUUGG ..((((((((((((((..(((.......)))....-..)))))))((..(((.((.(((((.((.((((...)))).))-.))))).)).)))..))..---.....))))))) ( -28.00, z-score = -0.91, R) >droEre2.scaffold_4690 7387317 101 + 18748788 -------GUACCGCA-AUGCUUUAUAGUGGUUUUG-UCGUGUGGUUUUAAGUUCCAGGAGAACGAGAAGAGGCUUUGUU-AUCUUCAGGCACUAGGAUA---UUAAAACCUUGG -------.(((.(((-(.(((.......))).)))-).))).((((((((..(((.((.(.....(((((.........-.)))))...).)).)))..---)))))))).... ( -21.60, z-score = 0.39, R) >droVir3.scaffold_12970 9771805 93 + 11907090 -----------AACUAAAUAACUGGAGUUGAAUUGCGGUUGUAGUUUCAAGUUCCAAGAGAGUAGGAAGAGUAG---UG---CUUCGAACACUAGGAUA---UUGAAACAGAA- -----------......(((((((.(((...))).))))))).(((((((..(((...((.((..((((.....---..---))))..)).)).)))..---)))))))....- ( -21.70, z-score = -1.56, R) >droMoj3.scaffold_6473 5146052 89 - 16943266 -----------AGCCAAAAUACUCGAUUUGUUUU-CAUUUGUAGUUUCAAGUUCCAAGAGAGUAAGAAGAGUUG---CG---CUUCAGACACUAGGAUA---UUGAAACA---- -----------.......................-........(((((((..(((...((.((..((((.....---..---))))..)).)).)))..---))))))).---- ( -16.20, z-score = -0.33, R) >droGri2.scaffold_14853 7080832 90 - 10151454 ----------------AUUAAUUAUGAUUAAUGUGCAAUUGUCGUUUCAAGUUCCAAGAGAGUAGGAAGAGGAG---UGUCGCUUCAAACACUAGGAUA---UUGAAACCAA-- ----------------.........(((.(((.....))))))(((((((..(((...((.((..((((.((..---..)).))))..)).)).)))..---)))))))...-- ( -16.80, z-score = -0.47, R) >droSim1.chrX 13221348 85 + 17042790 ---------------------GAAUGCUUUCUUUC-UAAUGUAGUCUUAAAUUCCAGGAGAGCGGGAAGUGGUUUAAU----CUUCAAGCACUAAUAUAA--UUAACACUUAU- ---------------------...(((((((((..-.(((..........)))..)))))))))...((((.((....----....)).)))).......--...........- ( -11.00, z-score = 0.96, R) >droAna3.scaffold_13248 2278755 97 + 4840945 --------------UUAAACAUUAUAAUAUUGAAGCUAUUGUGGCUUUAGUUUCCAUGAGAGCAAGAAGAGGCUAUAU---CUUUCAAGCACAAGGAUAAUAUUAAAACCUAUU --------------..........(((((((((((((.....))))))....(((.((...((..(((((.......)---))))...)).)).))).)))))))......... ( -14.70, z-score = 0.33, R) >dp4.chrXL_group1e 8801468 92 - 12523060 ------------------AUAUCCGAAUUGG-UUGCAGGUGUAGUUUUAAGUUCCAAGAGAGCAAGAUGAGGGGAUAUGUAUCGUCAAGCACUAGGAUA---UUAAAACUUGUG ------------------((((((....((.-...))(((((........((((.....))))..(((((...........)))))..))))).)))))---)........... ( -16.60, z-score = 0.32, R) >droPer1.super_17 1272345 92 - 1930428 ------------------AUAUCCGAAUUUG-UUGCAGGUGUAGUUUUAAGUUCCAAGAGAGCAAGAUGAGGGGAUAUGUAUCGUCAAGCACUAGGAUA---UUAAAACUUGUG ------------------(((((((((((((-..((.......))..)))))))....((.((..(((((...........)))))..)).)).)))))---)........... ( -18.20, z-score = -0.39, R) >consensus _____________C__AUACAUUAUAAUUGUUUUGCUAGUGUAGUUUUAAGUUCCAAGAGAGCAAGAAGAGGCUUUAUG_AUCUUCAAGCACUAGGAUA___UUAAAACCUAGG ..........................................((((((((..(((.((.(.....((((.............))))...).)).))).....)))))))).... ( -9.67 = -8.92 + -0.75)
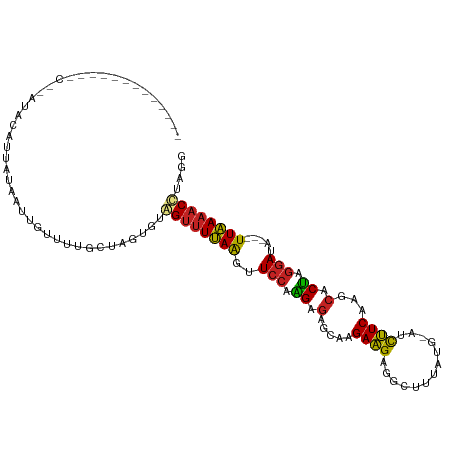
| Location | 17,040,609 – 17,040,718 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 61.91 |
| Shannon entropy | 0.75938 |
| G+C content | 0.37023 |
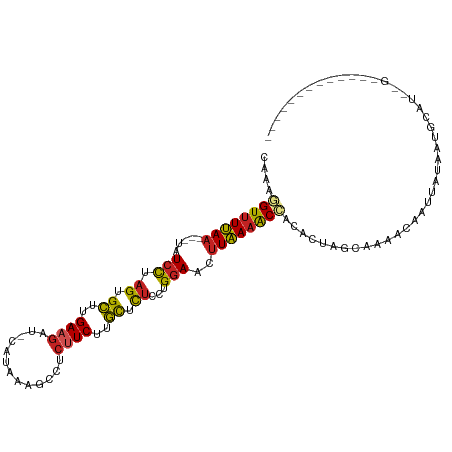
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -11.95 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17040609 109 - 22422827 ACAAGGUUUUAA---UGUCCUAGUGCUUGAAGAU-AUCAAAGCCUCUUCUUGUUCUCCGGGAACUUAAAACCACAUUA-CAAAACCAUUAUAAAGCAUAUGCGGUACCUUGGUU ....((((((((---..(((..(.((..(((((.-.........)))))..))...)..)))..))))))))......-...(((((...((..((....))..))...))))) ( -26.50, z-score = -2.35, R) >droSec1.super_17 881707 109 - 1527944 CCAAGGUUUUAA---UGUCCUAGUGCCUGAAGAU-AUAAAAUCCUCUUCUUGUUUUCCUGGAAUUUAAAACCACACUA-CAAAACCAUUAUAAAGCAUAUGCGGUACCUUGGUG ....((((((((---..(((..(.((..(((((.-.........)))))..))...)..)))..))))))))......-....((((...((..((....))..))...)))). ( -23.60, z-score = -2.13, R) >droYak2.chrX 15650330 109 - 21770863 CCAAGGUUUUAA---UAUCCUAGUGCCUGAAGAU-CACAAAGCCUCUUCUCGUUCUCCUGGAACUUAAAACCACACGA-CAAAACCAUUAUAAAGCAUAUGCGGUACCUUGGUG ....((((((((---..(((.((.((..(((((.-.........)))))..)).))...)))..))))))))......-....((((...((..((....))..))...)))). ( -23.20, z-score = -1.77, R) >droEre2.scaffold_4690 7387317 101 - 18748788 CCAAGGUUUUAA---UAUCCUAGUGCCUGAAGAU-AACAAAGCCUCUUCUCGUUCUCCUGGAACUUAAAACCACACGA-CAAAACCACUAUAAAGCAU-UGCGGUAC------- ....((((((((---..(((.((.((..(((((.-.........)))))..)).))...)))..))))))))......-....(((.(..........-.).)))..------- ( -18.50, z-score = -1.32, R) >droVir3.scaffold_12970 9771805 93 - 11907090 -UUCUGUUUCAA---UAUCCUAGUGUUCGAAG---CA---CUACUCUUCCUACUCUCUUGGAACUUGAAACUACAACCGCAAUUCAACUCCAGUUAUUUAGUU----------- -....(((((((---.....((((((.....)---))---)))...((((.........)))).)))))))................................----------- ( -13.80, z-score = -1.24, R) >droMoj3.scaffold_6473 5146052 89 + 16943266 ----UGUUUCAA---UAUCCUAGUGUCUGAAG---CG---CAACUCUUCUUACUCUCUUGGAACUUGAAACUACAAAUG-AAAACAAAUCGAGUAUUUUGGCU----------- ----.(((((((---..(((.((.((..((((---..---.....))))..))...)).)))..)))))))........-....((((........))))...----------- ( -15.40, z-score = -1.04, R) >droGri2.scaffold_14853 7080832 90 + 10151454 --UUGGUUUCAA---UAUCCUAGUGUUUGAAGCGACA---CUCCUCUUCCUACUCUCUUGGAACUUGAAACGACAAUUGCACAUUAAUCAUAAUUAAU---------------- --...(((((((---..(((.((.((..((((.(...---...).))))..))...)).)))..)))))))...........................---------------- ( -15.90, z-score = -1.87, R) >droSim1.chrX 13221348 85 - 17042790 -AUAAGUGUUAA--UUAUAUUAGUGCUUGAAG----AUUAAACCACUUCCCGCUCUCCUGGAAUUUAAGACUACAUUA-GAAAGAAAGCAUUC--------------------- -...(((.((((--...((..((.((..((((----.........))))..)).))..))....)))).)))......-..............--------------------- ( -9.30, z-score = 0.80, R) >droAna3.scaffold_13248 2278755 97 - 4840945 AAUAGGUUUUAAUAUUAUCCUUGUGCUUGAAAG---AUAUAGCCUCUUCUUGCUCUCAUGGAAACUAAAGCCACAAUAGCUUCAAUAUUAUAAUGUUUAA-------------- ....(((((((......(((.((.((..(((.(---(.......)))))..))...)).)))...)))))))............................-------------- ( -13.70, z-score = 0.35, R) >dp4.chrXL_group1e 8801468 92 + 12523060 CACAAGUUUUAA---UAUCCUAGUGCUUGACGAUACAUAUCCCCUCAUCUUGCUCUCUUGGAACUUAAAACUACACCUGCAA-CCAAUUCGGAUAU------------------ ....((((((((---..(((.((.((..((.((...........)).))..))...)).)))..))))))))..........-((.....))....------------------ ( -14.00, z-score = -1.70, R) >droPer1.super_17 1272345 92 + 1930428 CACAAGUUUUAA---UAUCCUAGUGCUUGACGAUACAUAUCCCCUCAUCUUGCUCUCUUGGAACUUAAAACUACACCUGCAA-CAAAUUCGGAUAU------------------ ....((((((((---..(((.((.((..((.((...........)).))..))...)).)))..))))))))..........-.............------------------ ( -13.20, z-score = -1.40, R) >consensus CAAAGGUUUUAA___UAUCCUAGUGCUUGAAGAU_CAUAAAGCCUCUUCUUGCUCUCCUGGAACUUAAAACCACACUAGCAAAACAAUUAUAAUGCAU__G_____________ ....((((((((.....(((.((.((..((((.............))))..)).))...)))..)))))))).......................................... (-11.95 = -11.35 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:38 2011