| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,040,020 – 17,040,082 |
| Length | 62 |
| Max. P | 0.997959 |

| Location | 17,040,020 – 17,040,082 |
|---|---|
| Length | 62 |
| Sequences | 12 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.51234 |
| G+C content | 0.41457 |
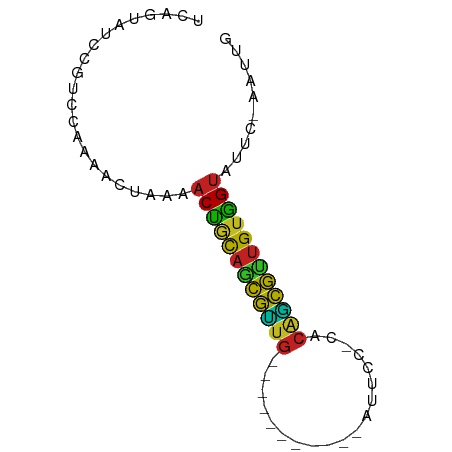
| Mean single sequence MFE | -14.65 |
| Consensus MFE | -13.51 |
| Energy contribution | -11.83 |
| Covariance contribution | -1.68 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.997959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
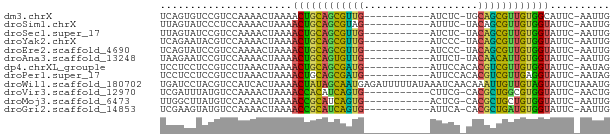
>dm3.chrX 17040020 62 - 22422827 UCAGUGUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCUC-UGCAGCGUUGUGGCAUUC-AAUUG ..((((((.((.....)).......(((((((((-----------.....-..))))))))))))))).-..... ( -13.60, z-score = -0.87, R) >droSim1.chrX 13221276 62 - 17042790 UUAGUAUCCCUCCAAAACUAAAACUGCAGCGUAG-----------AUUUC-UACAGCGUUGUGGUAUUC-AAUUG (((((...........))))).((..((((((((-----------....)-)...))))))..))....-..... ( -12.30, z-score = -2.04, R) >droSec1.super_17 881121 62 - 1527944 UUAGUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCUC-UACAGCGUUGUGGUAUUC-AAUUG (((((...........))))).((..((((((((-----------.....-..))))))))..))....-..... ( -15.80, z-score = -3.23, R) >droYak2.chrX 15649744 62 - 21770863 UCAGAAUACGUCCAAAACUAAAACUGCAGCGUUG-----------AUCCC-UACAGCGUUGUGGUAUUC-AAUUG ...((((((((.....)).......(((((((((-----------.....-..))))))))).))))))-..... ( -16.10, z-score = -3.09, R) >droEre2.scaffold_4690 7386757 62 - 18748788 UCAGUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCCC-UACAGCGUUGUGGUAUUC-AAUUG ......................((..((((((((-----------.....-..))))))))..))....-..... ( -15.10, z-score = -3.10, R) >droAna3.scaffold_13248 2278285 62 - 4840945 UAAGAAUCCGUCCAAAACUAAAACUGCAGUGUUG-----------AUUCU-UACAACAUUGUGGUAUUC-AAUUG ......................((..((((((((-----------.....-..))))))))..))....-..... ( -13.70, z-score = -2.80, R) >dp4.chrXL_group1e 8801046 63 + 12523060 UCCUCCUCCGUCCUAAACUAAAACUGCAGCGAUG-----------AUUCCACACGUCGUUGUGGUAUUC-AAUAG ......................((..((((((((-----------........))))))))..))....-..... ( -15.40, z-score = -3.74, R) >droPer1.super_17 1271923 63 + 1930428 UCCUCCUCCGUCCUAAACUAAAACUGCAGCGAUG-----------AUUCCACACGUCGUUGAGGUAUUC-AAUAG ......................(((.((((((((-----------........)))))))).)))....-..... ( -12.00, z-score = -1.97, R) >droWil1.scaffold_180702 1293287 75 + 4511350 UGAUCCUACGUCCAUCACUAAAACUAUAGCAAUGAGAUUUUUAUAAAUCAACAAAUUGUUGUAGUAUUCUAAAUG ......................(((((((((((..(((((....))))).....))))))))))).......... ( -12.90, z-score = -2.64, R) >droVir3.scaffold_12970 9771334 62 - 11907090 UCGAUUUAUGUCCAAAACUAAAACCACAUCAGUG-----------CUUCG-CACGCUGGCGUGGUAUUC-AACUG ..(((....)))..........(((((..(((((-----------.....-..)))))..)))))....-..... ( -14.30, z-score = -1.90, R) >droMoj3.scaffold_6473 5145813 62 + 16943266 UUGGCUUAUGUCCACAACUAAAACCGCAUCAGUG-----------ACUCG-CACGCUGCUGUGGUAUUC-AAUUG .((((....).)))........((((((.(((((-----------.....-..))))).))))))....-..... ( -15.80, z-score = -1.71, R) >droGri2.scaffold_14853 7080356 62 + 10151454 UCGAAGUAUGUCCAAAACUAAAACCGCAUCAGUG-----------AUUCA-CACGCUGAUGUGGUAUUC-AAUUG ......................((((((((((((-----------.....-..))))))))))))....-..... ( -18.80, z-score = -3.91, R) >consensus UCAGUAUCCGUCCAAAACUAAAACUGCAGCGUUG___________AUUCC_CACAGCGUUGUGGUAUUC_AAUUG ......................((((((((((((...................)))))))))))).......... (-13.51 = -11.83 + -1.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:36 2011