
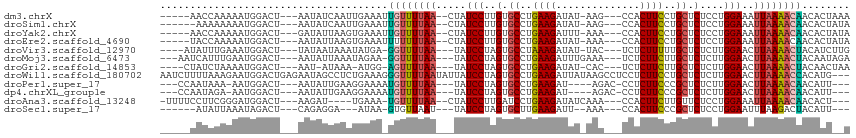
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,039,414 – 17,039,911 |
| Length | 497 |
| Max. P | 0.999620 |

| Location | 17,039,414 – 17,039,515 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.49 |
| Shannon entropy | 0.57182 |
| G+C content | 0.32790 |
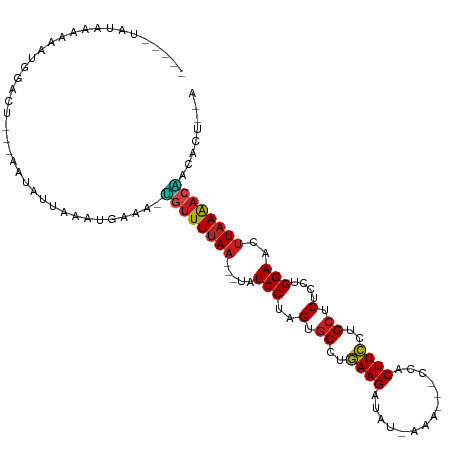
| Mean single sequence MFE | -16.26 |
| Consensus MFE | -10.29 |
| Energy contribution | -10.02 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
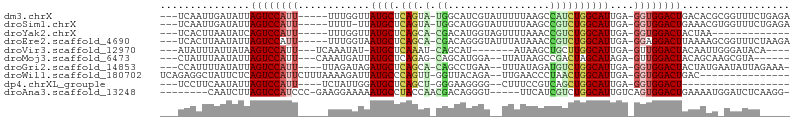
>dm3.chrX 17039414 101 - 22422827 -----AACCAAAAAUGGACU---AAUAUCAAUUGAAAUUGUUUUAA--CUAUCCUUGUGCCUGAAGAUAU-AAG---CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAAA -----..(((....)))...---..............(((((((((--...(((..(.((..((((....-...---...))))..)).)....)))..)))))))))....... ( -14.80, z-score = -1.92, R) >droSim1.chrX 13220673 100 - 17042790 ------AAAAAAAAUGGACU---AAUAUCAAUUGAAAUUGUUUUAA--CUAUCCUUGUGCCUGAAGAUAU-AAG---CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUA ------.......((((...---....((....))..(((((((((--...(((..(.((..((((....-...---...))))..)).)....)))..)))))))))..)))). ( -14.00, z-score = -1.70, R) >droYak2.chrX 15649482 101 - 21770863 -----AACCAAAAAUGGACU---GAUAUUAAGUGAAAUUGUUUUAA--CUAUCCUUGUGCCUGAAGAUUU-AAA---CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUA -----..(((....)))...---.......((((...(((((((((--...(((..(.((..((((....-...---...))))..)).)....)))..)))))))))))))... ( -17.70, z-score = -2.81, R) >droEre2.scaffold_4690 7386497 101 - 18748788 -----UACCAAAAAUGGACU---AAUAUUAAGUGAAAUUUUUUUAA--CUAUCCUUGUGCCUGAAGAUAU-AAA---CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUA -----..(((....)))...---.......((((...((.((((((--...(((..(.((..((((....-...---...))))..)).)....)))..)))))).))))))... ( -11.70, z-score = -0.94, R) >droVir3.scaffold_12970 9770711 100 - 11907090 ----AUAUUUGAAAUGGACU---UAUAAUAAAUAUGA-GGUUUUAA---UAUCCUAGUGCCUAAAGAUAU-UAC---UCUCUUUUUGCUCUCUUGGAACUUAAAACUACAUCUUG ----......((.(((..((---((((.....)))))-)(((((((---..(((.((.((..(((((...-...---..)))))..))...)).)))..)))))))..))).)). ( -17.50, z-score = -2.31, R) >droMoj3.scaffold_6473 5144973 102 + 16943266 ---AAUCAUUUGAAUGGACU---AAUAUUAAAUAGAA-GGUUUUAA---UAUCCUAGUGCCUGAAGAUUUGAAA---UCUCUUCUUGCUCUCUUGGAACUUAAAACUACAAUAGA ---..((((((((((.....---.)).)))))).)).-((((((((---..(((.((.((..(((((.......---..)))))..))...)).)))..))))))))........ ( -17.30, z-score = -0.64, R) >droGri2.scaffold_14853 7080069 98 + 10151454 ----CUAUCUAAAAUGGACU---AAU-AUAAA-AUGG-AGUUUUAA---UAUCCUAGUGCCUGAAGAUAU-CAC---UCUCUUCUUGCUCUCUUGGAACUUAAAACUACAACUAA ----................---...-.....-.((.-((((((((---..(((.((.((..(((((...-...---..)))))..))...)).)))..)))))))).))..... ( -18.50, z-score = -2.96, R) >droWil1.scaffold_180702 1292628 112 + 4511350 AAUCUUUUAAAGAAUGGACUGAGAAUAGCCUCUGAAAGGGUUUUAAUAUUAUCCUAGUGCCUGAAGAUUAUAAGCCUCCUCUUCCUGCUCUCUUGGAACUUAAAACCACAUG--- ...(((((..(((..((.(((....)))))))))))))((((((((.....(((.((.((..(((((............)))))..))...)).)))..)))))))).....--- ( -20.00, z-score = 0.03, R) >droPer1.super_17 1271305 97 + 1930428 ---CCAAUAAA-AAUGGACU---AAUAUUGAAGGAAAAUGUUUUAA---UAUCCUAGUGCCUGAAGAU----AGAC-CCUCUUCCCGCUCUCUUGGAACUUAAAACAACAUU--- ---(((.....-..)))...---...............((((((((---..(((.((.((..(((((.----....-..)))))..))...)).)))..)))))))).....--- ( -17.10, z-score = -1.62, R) >dp4.chrXL_group1e 8800418 97 + 12523060 ---CCAAUAGA-AAUGGACU---AAUAUUGAAGGAAAAUGUUUUAA---UAUCCUAGUGCCUGAAGAU----AGAC-CCUCUUCCCGCUCUCUUGGAACUUAAAACAACAUU--- ---.(((((..-........---..)))))........((((((((---..(((.((.((..(((((.----....-..)))))..))...)).)))..)))))))).....--- ( -17.40, z-score = -1.29, R) >droAna3.scaffold_13248 2277680 98 - 4840945 -UUUUCCUUCGGGAUGGACU---AAGAU----UGAAA-UGUUUUAA--CUAUCCUUGAUCCUGAAGAUAUCAAA---CCACUUCUUGUUCUCCUGGAAAUUAAAACAACACU--- -.((((((((((((((((.(---(((((----.....-.)))))).--...)))...))))))))((...(((.---.......))).))....))))).............--- ( -16.30, z-score = -0.33, R) >droSec1.super_17 881208 91 - 1527944 ------AUAUUAAAUAGACU---CAGAGGA---AUAA-GUGUUAAU---UAUCCUAGUGCUUGAAGAUU--AAA---CCACUUCCCGCUCUCCUGGAAUUUAAGACUACAUU--- ------..............---.......---...(-((.((((.---..(((.((.((..((((...--...---...))))..)).))...)))..)))).))).....--- ( -12.80, z-score = 0.30, R) >consensus _____UAUAAAAAAUGGACU___AAUAUUAAAUGAAA_UGUUUUAA___UAUCCUAGUGCCUGAAGAUAU_AAA___CCACUUCCUGCUCUCCUGGAACUUAAAACAACACU__A ......................................((((((((.....(((..(.((..((((..............))))..)).)....)))..))))))))........ (-10.29 = -10.02 + -0.27)
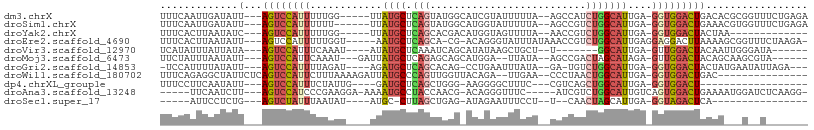
| Location | 17,039,486 – 17,039,583 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 54.15 |
| Shannon entropy | 0.95845 |
| G+C content | 0.39072 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -7.19 |
| Energy contribution | -7.68 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17039486 97 + 22422827 UUUCAAUUGAUAUU---AGUCCAUUUUUGG-----UUAUGCUCAGUAUGGCAUCGUAUUUUUA--AGCCAUCUGGCAUUGA-GGUGGACUGACACGCGGUUUCUGAGA (((((((((...((---((((((((((...-----..((((.(((.(((((............--.)))))))))))).))-))))))))))....))))...))))) ( -30.42, z-score = -3.04, R) >droSim1.chrX 13220745 96 + 17042790 UUUCAAUUGAUAUU---AGUCCAUUUUUU------UUAUGCUCAGUAUGGCAUGGUAUUUUUA--AGCCGUCUGGCAUUGA-GGUGGACUGAAACGUGGUUUCUGAGA (((((.........---((((((((((..------..((((.(((.(((((.(((.....)))--.)))))))))))).))-))))))))(((((...)))))))))) ( -30.90, z-score = -3.74, R) >droYak2.chrX 15649554 84 + 21770863 UUUCACUUAAUAUC---AGUCCAUUUUUGG-----UUAUGCUCAGCACGACAUGGUAGUUUUA--AACCGUCUGGCAUUGA-GGUGGACUACUAA------------- ..............---((((((((((...-----..((((.(((......(((((.......--.)))))))))))).))-)))))))).....------------- ( -21.00, z-score = -1.79, R) >droEre2.scaffold_4690 7386569 97 + 18748788 UUUCACUUAAUAUU---AGUCCAUUUUUGGU-----AAUGCUCAGCA-CG-ACAGGGUAUUUAUAAACCGUCUGGCAUUGAGGAGGACUUAAAAGCGGUUUCUAAGA- ............((---((.((.((((.(((-----..(.((((((.-.(-((.((...........)))))..))..)))).)..))).))))..))...))))..- ( -19.00, z-score = -0.07, R) >droVir3.scaffold_12970 9770781 85 + 11907090 UCAUAUUUAUUAUA---AGUCCAUUUCAAAU----AUAUGCUCAAAUCAGCAUAUAAGCUGCU--U-------GGCAUUGA-GUUGGACUACAAUUGGGAUA------ ..............---((((((.(((((.(----(((((((......)))))))).(((...--.-------))).))))-).))))))............------ ( -23.10, z-score = -3.06, R) >droMoj3.scaffold_6473 5145044 91 - 16943266 UUCUAUUUAAUAUU---AGUCCAUUCAAAU---GAUUAUGCUCAGAGCAGCAUGGA--UUAUA--AGCCGACUAGCAUAGA-GUUGGACUACAGCAAGCGUA------ .............(---(((((((((..((---(((((((((......))))).))--)))).--.((......))...))-).)))))))...........------ ( -19.30, z-score = -0.74, R) >droGri2.scaffold_14853 7080138 92 - 10151454 -UCCAUUUUAUAUU---AGUCCAUUUUAGAU----AGAUGCUCAGCACAG-CCUGAAUUUAUA--GA-UGUCUGGCAUUGA-GGUGGACUACUAUGAAUAUUAGA--- -.....((((((.(---((((((((((....----.(((((.(((.(((.-.(((......))--).-)))))))))))))-))))))))).)))))).......--- ( -27.00, z-score = -3.10, R) >droWil1.scaffold_180702 1292703 88 - 4511350 UUUCAGAGGCUAUUCUCAGUCCAUUCUUUAAAAGAUUAUGCCCAGUUGGUUACAGA--UUGAA--CCCUAACUGGCAUUGA-GGUGGACUGAC--------------- ...............(((((((((.(((.........(((((.((((((((.....--...))--))..))))))))).))-)))))))))).--------------- ( -25.20, z-score = -1.86, R) >dp4.chrXL_group1e 8800485 78 - 12523060 UUUCCUUCAAUAUU---AGUCCAUUUCUAUUG----GAUGCUCAGCUGGG-AAGGGGCUUUC---CGUCAGCUGGCAUUGA-GGUGGACU------------------ ..............---((((((((((.....----(((((.((((((((-(((....))))---)..)))))))))))))-))))))))------------------ ( -31.30, z-score = -3.33, R) >droAna3.scaffold_13248 2277750 92 + 4840945 -----UUCAAUCUU---AGUCCAUCCCGAAGGA-AAAAUGCCUACCAACG-ACAGGGUUUC-----AUCGUCUGGCAUUGUCAGUGGACUGAAAAUGGAUCUCAAGG- -----((((.(.((---(((((((((....)))-..((((((.....(((-(.........-----.))))..)))))).....)))))))).).))))........- ( -21.60, z-score = -0.12, R) >droSec1.super_17 881276 73 + 1527944 -----AUUCCUCUG---AGUCUAUUUAAUAU----AUGC-CUUAGCUGAG-AUAGAAUUUCCU--U--CAACUAGCAUUGA-GGUAGACUCA---------------- -----........(---(((((((((.....----((((-..(((.((((-...........)--)--)).)))))))..)-))))))))).---------------- ( -18.00, z-score = -1.99, R) >consensus UUUCAUUUAAUAUU___AGUCCAUUUUUAAU____UUAUGCUCAGCACGG_AUAGAAUUUUUA__AACCGUCUGGCAUUGA_GGUGGACUAAAA___G__U_______ .................((((((((............((((.(((..........................)))))))....)))))))).................. ( -7.19 = -7.68 + 0.49)
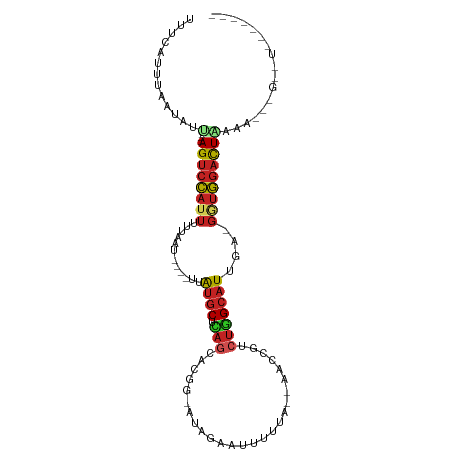
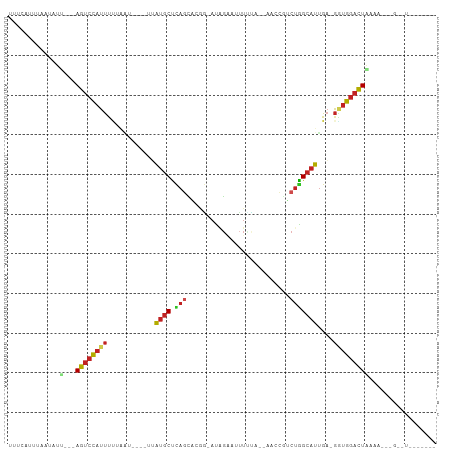
| Location | 17,039,486 – 17,039,583 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 54.15 |
| Shannon entropy | 0.95845 |
| G+C content | 0.39072 |
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -4.78 |
| Energy contribution | -5.07 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17039486 97 - 22422827 UCUCAGAAACCGCGUGUCAGUCCACC-UCAAUGCCAGAUGGCU--UAAAAAUACGAUGCCAUACUGAGCAUAA-----CCAAAAAUGGACU---AAUAUCAAUUGAAA .............((((.((((((..-...((((((((((((.--............))))).))).))))..-----.......))))))---.))))......... ( -21.86, z-score = -2.76, R) >droSim1.chrX 13220745 96 - 17042790 UCUCAGAAACCACGUUUCAGUCCACC-UCAAUGCCAGACGGCU--UAAAAAUACCAUGCCAUACUGAGCAUAA------AAAAAAUGGACU---AAUAUCAAUUGAAA ..((((((((...)))))((((((..-...(((((((..(((.--............)))...))).))))..------......))))))---.........))).. ( -20.24, z-score = -3.44, R) >droYak2.chrX 15649554 84 - 21770863 -------------UUAGUAGUCCACC-UCAAUGCCAGACGGUU--UAAAACUACCAUGUCGUGCUGAGCAUAA-----CCAAAAAUGGACU---GAUAUUAAGUGAAA -------------....(((((((..-...(((((((((((..--.............)))).))).))))..-----.......))))))---)............. ( -15.80, z-score = -0.82, R) >droEre2.scaffold_4690 7386569 97 - 18748788 -UCUUAGAAACCGCUUUUAAGUCCUCCUCAAUGCCAGACGGUUUAUAAAUACCCUGU-CG-UGCUGAGCAUU-----ACCAAAAAUGGACU---AAUAUUAAGUGAAA -..........(((((...(((((.....((((((((((((...............)-))-).))).)))))-----.........)))))---......)))))... ( -18.20, z-score = -1.45, R) >droVir3.scaffold_12970 9770781 85 - 11907090 ------UAUCCCAAUUGUAGUCCAAC-UCAAUGCC-------A--AGCAGCUUAUAUGCUGAUUUGAGCAUAU----AUUUGAAAUGGACU---UAUAAUAAAUAUGA ------.......(((((((((((..-((((((((-------(--((((((......)))).)))).))))..----...)))..))))).---))))))........ ( -24.00, z-score = -4.05, R) >droMoj3.scaffold_6473 5145044 91 + 16943266 ------UACGCUUGCUGUAGUCCAAC-UCUAUGCUAGUCGGCU--UAUAA--UCCAUGCUGCUCUGAGCAUAAUC---AUUUGAAUGGACU---AAUAUUAAAUAGAA ------...........(((((((..-..((((((((.((((.--.....--.....))))..)).)))))).((---....)).))))))---)............. ( -20.10, z-score = -1.27, R) >droGri2.scaffold_14853 7080138 92 + 10151454 ---UCUAAUAUUCAUAGUAGUCCACC-UCAAUGCCAGACA-UC--UAUAAAUUCAGG-CUGUGCUGAGCAUCU----AUCUAAAAUGGACU---AAUAUAAAAUGGA- ---..........(((.(((((((..-...((((((((((-((--(........)))-.))).))).))))..----........))))))---).)))........- ( -16.86, z-score = -0.67, R) >droWil1.scaffold_180702 1292703 88 + 4511350 ---------------GUCAGUCCACC-UCAAUGCCAGUUAGGG--UUCAA--UCUGUAACCAACUGGGCAUAAUCUUUUAAAGAAUGGACUGAGAAUAGCCUCUGAAA ---------------.((((((((..-...((((((((...((--(((..--...).)))).))).)))))..(((.....))).))))))))............... ( -24.50, z-score = -2.02, R) >dp4.chrXL_group1e 8800485 78 + 12523060 ------------------AGUCCACC-UCAAUGCCAGCUGACG---GAAAGCCCCUU-CCCAGCUGAGCAUC----CAAUAGAAAUGGACU---AAUAUUGAAGGAAA ------------------((((((..-((.((((((((((..(---(((......))-)))))))).)))).----.....))..))))))---.............. ( -23.70, z-score = -3.34, R) >droAna3.scaffold_13248 2277750 92 - 4840945 -CCUUGAGAUCCAUUUUCAGUCCACUGACAAUGCCAGACGAU-----GAAACCCUGU-CGUUGGUAGGCAUUUU-UCCUUCGGGAUGGACU---AAGAUUGAA----- -........((.(((((.((((((.....((((((.((((((-----........))-))))....))))))..-(((....)))))))))---))))).)).----- ( -28.90, z-score = -2.28, R) >droSec1.super_17 881276 73 - 1527944 ----------------UGAGUCUACC-UCAAUGCUAGUUG--A--AGGAAAUUCUAU-CUCAGCUAAG-GCAU----AUAUUAAAUAGACU---CAGAGGAAU----- ----------------((((((((..-...((((((((((--(--.((.....))..-.))))))..)-))))----........))))))---)).......----- ( -18.32, z-score = -1.88, R) >consensus _______A__C___UUUUAGUCCACC_UCAAUGCCAGACGGCU__UAAAAAUUCUAU_CCGUGCUGAGCAUAA____AUCAAAAAUGGACU___AAUAUUAAAUGAAA ..................((((((......(((((((..........................))).))))..............))))))................. ( -4.78 = -5.07 + 0.28)
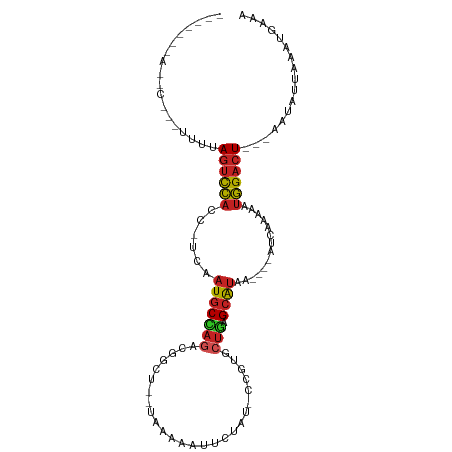
| Location | 17,039,488 – 17,039,583 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.88140 |
| G+C content | 0.40129 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
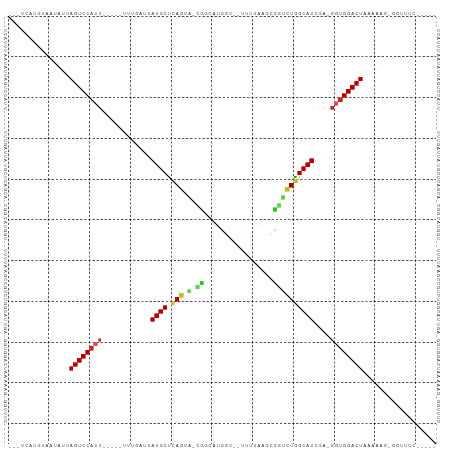
>dm3.chrX 17039488 95 + 22422827 ---UCAAUUGAUAUUAGUCCAUU-----UUUGGUUAUGCUCAGUA-UGGCAUCGUAUUUUUAAGCCAUCUGGCAUUGA-GGUGGACUGACACGCGGUUUCUGAGA ---..(((((...((((((((((-----((.....((((.(((.(-((((.............)))))))))))).))-))))))))))....)))))....... ( -29.52, z-score = -2.65, R) >droSim1.chrX 13220747 94 + 17042790 ---UCAAUUGAUAUUAGUCCAUU-----UUUU-UUAUGCUCAGUA-UGGCAUGGUAUUUUUAAGCCGUCUGGCAUUGA-GGUGGACUGAAACGUGGUUUCUGAGA ---..........((((((((((-----((..-..((((.(((.(-((((.(((.....))).)))))))))))).))-))))))))))................ ( -29.80, z-score = -3.31, R) >droYak2.chrX 15649556 82 + 21770863 ---UCACUUAAUAUCAGUCCAUU-----UUUGGUUAUGCUCAGCA-CGACAUGGUAGUUUUAAACCGUCUGGCAUUGA-GGUGGACUACUAA------------- ---............((((((((-----((.....((((.(((..-....(((((........)))))))))))).))-)))))))).....------------- ( -21.00, z-score = -1.70, R) >droEre2.scaffold_4690 7386571 95 + 18748788 ---UCACUUAAUAUUAGUCCAUU-----UUUGGUAAUGCUCAGCA-CGACAGGGUAUUUAUAAACCGUCUGGCAUUGA-GGAGGACUUAAAAGCGGUUUCUAAGA ---..........((((.((.((-----((.(((..(.((((((.-.(((.((...........)))))..))..)))-).)..))).))))..))...)))).. ( -19.00, z-score = -0.12, R) >droVir3.scaffold_12970 9770783 85 + 11907090 ---AUAUUUAUUAUAAGUCCAUU---UCAAAUAU-AUGCUCAAAU-CAGCAU-------AUAAGCUGCUUGGCAUUGA-GUUGGACUACAAUUGGGAUACA---- ---.(((((......((((((.(---((((.(((-(((((.....-.)))))-------))).(((....))).))))-).)))))).......)))))..---- ( -23.52, z-score = -3.29, R) >droMoj3.scaffold_6473 5145046 89 - 16943266 ---CUAUUUAAUAUUAGUCCAUU---CAAAUGAUUAUGCUCAGAG-CAGCAUGGA--UUAUAAGCCGACUAGCAUAGA-GUUGGACUACAGCAAGCGUA------ ---...........(((((((((---(..(((((((((((.....-.))))).))--))))..((......))...))-).)))))))...........------ ( -19.30, z-score = -0.82, R) >droGri2.scaffold_14853 7080139 93 - 10151454 ---CCAUUUUAUAUUAGUCCAUU----UUAGAUAGAUGCUCAGCA-CAGCCUGAA--UUUAUAGAUGUCUGGCAUUGA-GGUGGACUACUAUGAAUAUUAGAAA- ---....((((((.(((((((((----((.....(((((.(((.(-((..(((..--....))).)))))))))))))-))))))))).)))))).........- ( -27.00, z-score = -3.31, R) >droWil1.scaffold_180702 1292705 86 - 4511350 UCAGAGGCUAUUCUCAGUCCAUUCUUUAAAAGAUUAUGCCCAGUU-GGUUACAGA--UUGAACCCUAACUGGCAUUGA-GGUGGACUGAC--------------- .............(((((((((.(((.........(((((.((((-((((.....--...))))..))))))))).))-)))))))))).--------------- ( -25.20, z-score = -1.87, R) >dp4.chrXL_group1e 8800487 76 - 12523060 ---UCCUUCAAUAUUAGUCCAUU----UCUAUUGGAUGCUCAGCU-GGGAAGGGG--CUUUCCGUCAGCUGGCAUUGA-GGUGGACU------------------ ---............((((((((----((.....(((((.(((((-((((((...--.)))))..)))))))))))))-))))))))------------------ ( -31.30, z-score = -3.31, R) >droAna3.scaffold_13248 2277752 90 + 4840945 --------CAAUCUUAGUCCAUCCC-GAAGGAAAAAUGCCUACCAACGACAGGGU-----UUCAUCGUCUGGCAUUGUCAGUGGACUGAAAAUGGAUCUCAAGG- --------.....(((((((((((.-...)))..((((((.....((((......-----....))))..)))))).....))))))))...............- ( -20.40, z-score = 0.11, R) >consensus ___UCAUUUAAUAUUAGUCCAUU_____UUUGAUUAUGCUCAGCA_CGGCAUGGU__UUUUAAGCCGUCUGGCAUUGA_GGUGGACUAAAAAG_GGUUUC_____ ...............((((((((............((((.(((.........................)))))))....)))))))).................. ( -9.46 = -9.82 + 0.36)
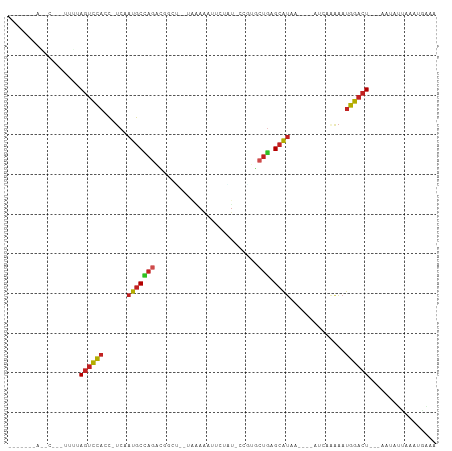
| Location | 17,039,488 – 17,039,583 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.88140 |
| G+C content | 0.40129 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.91 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
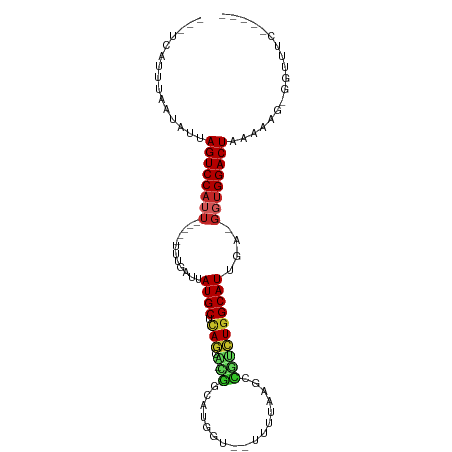
>dm3.chrX 17039488 95 - 22422827 UCUCAGAAACCGCGUGUCAGUCCACC-UCAAUGCCAGAUGGCUUAAAAAUACGAUGCCA-UACUGAGCAUAACCAAA-----AAUGGACUAAUAUCAAUUGA--- .............((((.((((((..-...((((((((((((.............))))-).))).)))).......-----..)))))).)))).......--- ( -21.86, z-score = -2.67, R) >droSim1.chrX 13220747 94 - 17042790 UCUCAGAAACCACGUUUCAGUCCACC-UCAAUGCCAGACGGCUUAAAAAUACCAUGCCA-UACUGAGCAUAA-AAAA-----AAUGGACUAAUAUCAAUUGA--- .....(((((...)))))((((((..-...(((((((..(((.............))).-..))).))))..-....-----..))))))............--- ( -19.44, z-score = -2.99, R) >droYak2.chrX 15649556 82 - 21770863 -------------UUAGUAGUCCACC-UCAAUGCCAGACGGUUUAAAACUACCAUGUCG-UGCUGAGCAUAACCAAA-----AAUGGACUGAUAUUAAGUGA--- -------------....(((((((..-...(((((((((((...............)))-).))).)))).......-----..)))))))...........--- ( -15.80, z-score = -0.67, R) >droEre2.scaffold_4690 7386571 95 - 18748788 UCUUAGAAACCGCUUUUAAGUCCUCC-UCAAUGCCAGACGGUUUAUAAAUACCCUGUCG-UGCUGAGCAUUACCAAA-----AAUGGACUAAUAUUAAGUGA--- ..........(((((...(((((...-..((((((((((((...............)))-).))).)))))......-----...)))))......))))).--- ( -18.20, z-score = -1.39, R) >droVir3.scaffold_12970 9770783 85 - 11907090 ----UGUAUCCCAAUUGUAGUCCAAC-UCAAUGCCAAGCAGCUUAU-------AUGCUG-AUUUGAGCAU-AUAUUUGA---AAUGGACUUAUAAUAAAUAU--- ----.........(((((((((((..-(((((((((((((((....-------..))))-.)))).))))-.....)))---..))))).))))))......--- ( -24.00, z-score = -3.99, R) >droMoj3.scaffold_6473 5145046 89 + 16943266 ------UACGCUUGCUGUAGUCCAAC-UCUAUGCUAGUCGGCUUAUAA--UCCAUGCUG-CUCUGAGCAUAAUCAUUUG---AAUGGACUAAUAUUAAAUAG--- ------...........(((((((..-..((((((((.((((......--.....))))-..)).)))))).((....)---).)))))))...........--- ( -20.10, z-score = -1.52, R) >droGri2.scaffold_14853 7080139 93 + 10151454 -UUUCUAAUAUUCAUAGUAGUCCACC-UCAAUGCCAGACAUCUAUAAA--UUCAGGCUG-UGCUGAGCAUCUAUCUAA----AAUGGACUAAUAUAAAAUGG--- -............(((.(((((((..-...(((((((((((((.....--...))).))-).))).))))........----..))))))).))).......--- ( -16.86, z-score = -1.05, R) >droWil1.scaffold_180702 1292705 86 + 4511350 ---------------GUCAGUCCACC-UCAAUGCCAGUUAGGGUUCAA--UCUGUAACC-AACUGGGCAUAAUCUUUUAAAGAAUGGACUGAGAAUAGCCUCUGA ---------------.((((((((..-...((((((((...(((((..--...).))))-.))).)))))..(((.....))).))))))))............. ( -24.50, z-score = -1.86, R) >dp4.chrXL_group1e 8800487 76 + 12523060 ------------------AGUCCACC-UCAAUGCCAGCUGACGGAAAG--CCCCUUCCC-AGCUGAGCAUCCAAUAGA----AAUGGACUAAUAUUGAAGGA--- ------------------((((((..-((.((((((((((..((((..--....)))))-))))).))))......))----..))))))............--- ( -23.70, z-score = -3.23, R) >droAna3.scaffold_13248 2277752 90 - 4840945 -CCUUGAGAUCCAUUUUCAGUCCACUGACAAUGCCAGACGAUGAA-----ACCCUGUCGUUGGUAGGCAUUUUUCCUUC-GGGAUGGACUAAGAUUG-------- -.(((((((.....))))((((((.....((((((.((((((...-----.....))))))....))))))..(((...-.))))))))))))....-------- ( -27.80, z-score = -2.29, R) >consensus _____GAAACC_CGUUUCAGUCCACC_UCAAUGCCAGACGGCUUAAAA__ACCAUGCCG_UGCUGAGCAUAAUCAAA_____AAUGGACUAAUAUUAAAUGA___ ..................((((((......(((((((.((((.............))))...))).))))..............))))))............... ( -9.72 = -9.91 + 0.19)

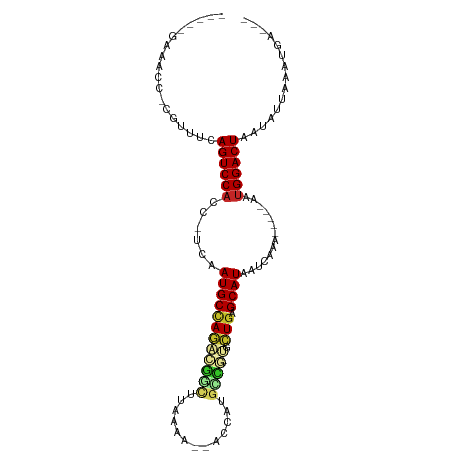
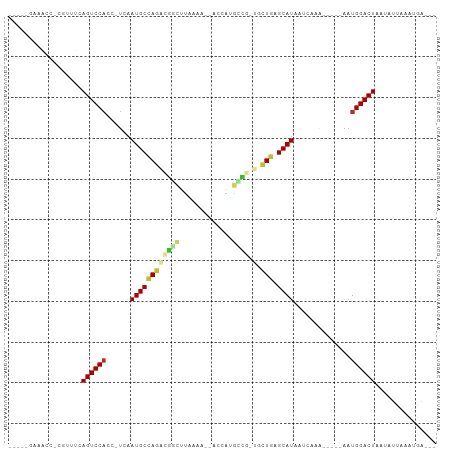
| Location | 17,039,583 – 17,039,642 |
|---|---|
| Length | 59 |
| Sequences | 12 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Shannon entropy | 0.49765 |
| G+C content | 0.33808 |
| Mean single sequence MFE | -7.67 |
| Consensus MFE | -6.09 |
| Energy contribution | -5.81 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883298 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 17039583 59 - 22422827 CUUAAGGGCAUGGCCUCAAAAAUGC-AACCUUAAUAAAG-C-AUAAAUCAACAAAAACCUAU .((((((((((..........))))-..)))))).....-.-.................... ( -7.40, z-score = -0.58, R) >droSim1.chrX 13220841 59 - 17042790 CUUAAGGGCAUGGCCUCAAAAAUGA-AACCUUAAUAAAG-C-AUAAAUCAACAAAAACCUAA .((((((.(((..........))).-..)))))).....-.-.................... ( -6.30, z-score = -0.43, R) >droYak2.chrX 15649305 59 - 21770863 CUUAAGGGCAUGGGCUCAAAAGUGC-AACCUUAAUAAAG-CAAUAAAUCAACAAAACCUCC- .((((((...((.(((....))).)-).)))))).....-.....................- ( -8.70, z-score = -0.42, R) >droEre2.scaffold_4690 7386666 59 - 18748788 CUUAAGGGCAUCGGCUUAAAAAUGC-AACCUUAAUAAAG-C-AUAAAUCAACAAUAACCCAU .((((((......((........))-..)))))).....-.-.................... ( -6.40, z-score = -0.33, R) >droPer1.super_17 1271473 53 + 1930428 CUUAAGGGCAUUGGCUUGAAUAUGCGAACCUUAAUAAAG-UUUAAAAUCAACAA-------- .((((((...((.((........)).)))))))).....-..............-------- ( -6.70, z-score = -0.25, R) >dp4.chrXL_group1e 8800590 60 + 12523060 CUUAAGGGCAUUGGCUUGAAUAUGCGAACCUUAAUAAAG-UUUAAAAUCAACAUCCACCCA- .((((((...((.((........)).)))))))).....-.....................- ( -6.70, z-score = 0.58, R) >droSec1.super_17 881033 56 - 1527944 CUUAAGGGCAUGGCCUCAAAAAUGC-AACCUUAAUAAAG-CAAUAAAUCAACAAAACC---- .((((((((((..........))))-..)))))).....-..................---- ( -7.40, z-score = -0.73, R) >droAna3.scaffold_13248 2277864 59 - 4840945 CUUAAGGGCAUUGCCUCAAAAAUGCGAACCUUAAUAAAG--UCUAAAUCAACAAUAAAAUA- .(((((((((((........)))))...)))))).....--....................- ( -8.80, z-score = -2.01, R) >droWil1.scaffold_180702 1292825 59 + 4511350 CUUAAGGAUAUCGUCUCGAAGAUAU-AACCUUAAUAAAGUUCAAAAAUCAACAAAAUCAU-- .(((((((((((........)))))-..))))))..........................-- ( -7.90, z-score = -2.46, R) >droGri2.scaffold_14853 7080273 51 + 10151454 CUUAAGGGCAUUGGCUAAAAGAUGCGAACCUUAAUAAAG--UCAAAAUCAACA--------- .(((((((((((........)))))...)))))).....--............--------- ( -8.60, z-score = -1.40, R) >droMoj3.scaffold_6473 5145168 51 + 16943266 CUUAAGGGCAUUGGCUAAAAGAUGCAUUCCUUAAUAAAG--UCAAAAUCAACA--------- .(((((((((((........)))))...)))))).....--............--------- ( -8.60, z-score = -1.30, R) >droVir3.scaffold_12970 9770903 50 - 11907090 -UUAAGGGCAUUGGCUCGAAGAUGCAAACCUUAAUAAAG--UCAAAAUCAACA--------- -(((((((((((........)))))...)))))).....--............--------- ( -8.50, z-score = -1.42, R) >consensus CUUAAGGGCAUUGGCUCAAAAAUGC_AACCUUAAUAAAG_CUAAAAAUCAACAAAAAC____ .((((((((((..........))))...))))))............................ ( -6.09 = -5.81 + -0.28)
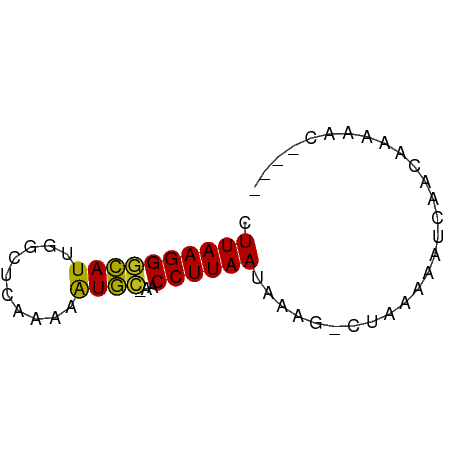
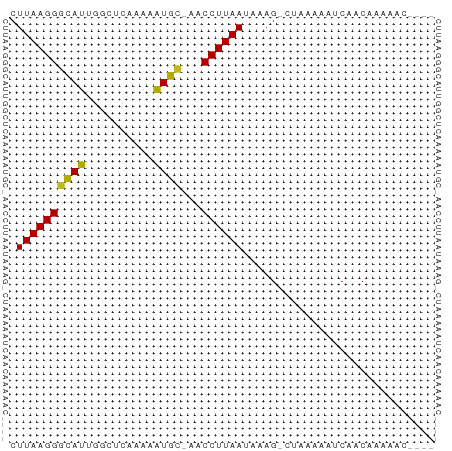
| Location | 17,039,642 – 17,039,753 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 68.49 |
| Shannon entropy | 0.60200 |
| G+C content | 0.40626 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -21.02 |
| Energy contribution | -18.90 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
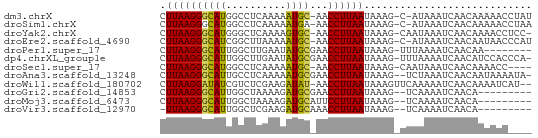
>dm3.chrX 17039642 111 - 22422827 --------AUUAUUCACCAAG-----GGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCCU-UACAGGGUUGUGGUAUUC-AACUGGACCAUAAUAAACCCCAUGAUUAGCUAUUCCU --------.(((.(((....(-----(((...(((((..(((((........((..((((.(((-----------.....-..))).))))..))....-...))))).)))))..))))..))).)))........ ( -23.86, z-score = -0.87, R) >droSim1.chrX 13220900 111 - 17042790 --------AUUAUUCACCAAG-----AGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCUC-UACAGCGUUGUGGUAUUC-AACUGGACCACAAUAAACCCCAUGAUUAGCUAUUCCU --------..(((((.....)-----)))).........(((((........((..((((((((-----------.....-..))))))))..))....-...)))))............................. ( -22.76, z-score = -2.39, R) >droYak2.chrX 15649364 111 - 21770863 --------AUUGUUCACCAAG-----AGUAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGCUG-----------AUUCC-CACAGCGUUGUGGUAUUC-AACUGGACUACAACAAACCCCAUGAUUAGCUAUUCCU --------.(((((......(-----..((....))..)(((((........((..((((((((-----------.....-..))))))))..))....-...)))))...)))))..................... ( -24.96, z-score = -2.61, R) >droEre2.scaffold_4690 7386725 95 - 18748788 -----------------------------UUCAGUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCCC-UACAGCGUUGUGGUAUUC-AAUUGGACCACAGGAAACCCCAUGGUUAGCUAUUCCU -----------------------------...(((.(((((((((.......((..((((((((-----------.....-..))))))))..))....-..))))))....)))((((....)))).)))...... ( -26.62, z-score = -2.75, R) >droPer1.super_17 1271526 95 + 1930428 ------------------------------UCCUCCUCCGUCCUAAACUAAAACUGCAGCGAUG-----------AUUCCACACGUCGUUGUGGUAUUC-AAUAGGACCAAACCAAAGCCCAUGAUUUGCUAUUCCU ------------------------------.........((((((.......((..((((((((-----------........))))))))..))....-..))))))........(((.........)))...... ( -23.02, z-score = -3.48, R) >dp4.chrXL_group1e 8800650 125 + 12523060 CCCAUCAAAAGAUCCACGAGAUGGUUAUCCUCCUCCCCCGUCCUAAACUAAAACUGCAGCGAUG-----------AUUCCACACGUCGUUGUGGUAUUC-AAUAGGACCAAACCAAAGCCCAUGAUUUGCUAUUCCU .........(((((...(((..((....))..)))....((((((.......((..((((((((-----------........))))))))..))....-..))))))...............)))))......... ( -29.22, z-score = -3.00, R) >droSec1.super_17 881089 111 - 1527944 --------AGCAUUCAUUAGA-----GAGAUUAGUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCUC-UACAGCGUUGUGGUAUUC-AAUUGGACCACAGGAAACCCCAUGGUUAGCUAUUCCU --------............(-----(.((.((((.(((((((((.......((..((((((((-----------.....-..))))))))..))....-..))))))....)))((((....)))).)))).)))) ( -30.12, z-score = -2.45, R) >droAna3.scaffold_13248 2277923 105 - 4840945 ------------CCCAUUUGU-----A--AACGAUAUCCGUCCAAAACUAAAACUGCAGCGUUG-----------AUCCU-CACAGCGUUGUGGUAUUC-AAUUGGACCAAAAUGAACCCCAUGGUUAACUAUUCCU ------------..((((((.-----.--..).......((((((.......((..((((((((-----------.....-..))))))))..))....-..))))))..)))))((((....)))).......... ( -24.42, z-score = -2.53, R) >droWil1.scaffold_180702 1292884 111 + 4511350 ----------------UAAA------UUCAAUUGAUUACGUCCAUCACUAAAACUAUAGCAAUGAGAUUUUCAUAAAUCAAUAAAUUGUUGUAGUAUUCUAAAUGGACUACAACAAAC----UGAUAAACUACUCCU ----------------....------.(((.(((.....((((((.......(((((((((((..(((((....))))).....))))))))))).......)))))).....)))..----)))............ ( -20.84, z-score = -3.66, R) >droVir3.scaffold_12970 9770954 107 - 11907090 ----------UCUUUUUGAA------CAC-UCGAUUUAUGUCCAAAACUAAAACCGCAUCAGUG-----------AUUUA-CACGCUGGCGUGGUAUUC-AAUUGGACUACAUAAAAGCCCAUGAUUAACUAUUCCU ----------.....((((.------...-)))).....((((((.......(((((..(((((-----------.....-..)))))..)))))....-..))))))............................. ( -21.02, z-score = -1.83, R) >droMoj3.scaffold_6473 5145219 111 + 16943266 ------UCAUUAUCUAUCAG------AGA-UUGGCUUAUGUCCACAACUAAAACCGCAUCAGUG-----------ACCCA-CACGCUGGUGUGGUAUUC-AAUUGGACUACAUUGAACCCCAUGGUUAACUAUUCCU ------............((------.((-.(((.....(((((........((((((((((((-----------.....-..))))))))))))....-...))))).......((((....))))..))).)))) ( -28.26, z-score = -1.88, R) >droGri2.scaffold_14853 7080324 108 + 10151454 ----------UCUGUAUAGA------AACUUCGAAGUAUGUCCAAAACUAAAACCGCAUCAGUG-----------AUUCA-CACGCUGAUGUGGUAUUC-AAUUGGACUACAUCAAAGCCCAUGAUUUACUAUUCCU ----------..........------..(((.((.(((.((((((.......((((((((((((-----------.....-..))))))))))))....-..))))))))).)).)))................... ( -30.42, z-score = -4.42, R) >consensus __________UAUUCAUCAA______AGCAUCAUUAUCCGUCCAAAACUAAAACUGCAGCGUUG___________AUCCC_CACAGCGUUGUGGUAUUC_AAUUGGACCACAACAAACCCCAUGAUUAGCUAUUCCU .......................................(((((........((((((((((((...................))))))))))))........)))))............................. (-21.02 = -18.90 + -2.12)
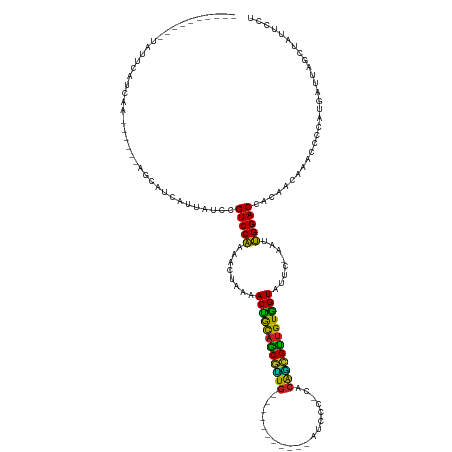
| Location | 17,039,753 – 17,039,911 |
|---|---|
| Length | 158 |
| Sequences | 12 |
| Columns | 182 |
| Reading direction | forward |
| Mean pairwise identity | 65.24 |
| Shannon entropy | 0.71997 |
| G+C content | 0.39435 |
| Mean single sequence MFE | -43.11 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.18 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
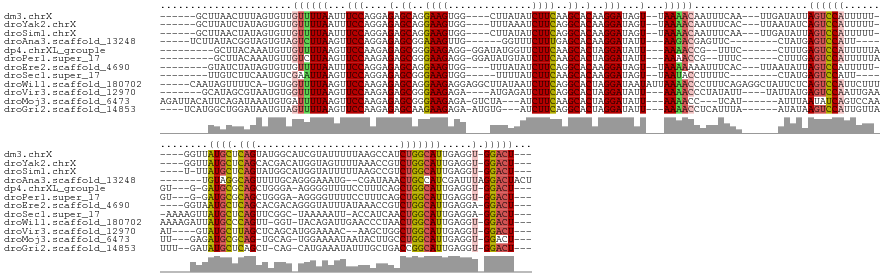
>dm3.chrX 17039753 158 + 22422827 ------GCUUAACUUUAGUGUUGUUUUAAUUUCCAGGAGAGCAGGAAGUGG----CUUAUAUCUUCAAGCACAAGGAUAGU--UAAAACAAUUUCAA---UUGAUAUUAGUCCAUUUUU-----GGUUAUGCUCAGUAUGGCAUCGUAUUUUUAAGCCAUCUGGCAUUGAGGU-GGACU--- ------...(((..((((((((((((((((((((..(...((..((((...----.......))))..)).)..))).)))--)))))))))....)---))))..)))(((((((((.-----....((((.(((.(((((.............)))))))))))).)))))-)))).--- ( -42.52, z-score = -2.19, R) >droYak2.chrX 15649475 158 + 21770863 ------GCUUAUCUAUAGUGUUGUUUUAAUUUCCAGGAGAGCAGGAAGUGG----UUUAAAUCUUCAGGCACAAGGAUAGU--UAAAACAAUUUCAC---UUAAUAUCAGUCCAUUUUU-----GGUUAUGCUCAGCACGACAUGGUAGUUUUAAACCGUCUGGCAUUGAGGU-GGACU--- ------..........((((((((((((((((((..(...((..((((...----.......))))..)).)..))).)))--))))))))...)))---).......((((((((((.-----....((((.(((......(((((........)))))))))))).)))))-)))))--- ( -39.20, z-score = -1.24, R) >droSim1.chrX 13221011 157 + 17042790 ------GCUUAACUAUAGUGUUGUUUUAAUUUCCAGGAGAGCAGGAAGUGG----CUUAUAUCUUCAGGCACAAGGAUAGU--UAAAACAAUUUCAA---UUGAUAUUAGUCCAUUUUU-----U-UUAUGCUCAGUAUGGCAUGGUAUUUUUAAGCCGUCUGGCAUUGAGGU-GGACU--- ------...........(.(((((((((((((((..(...((..((((...----.......))))..)).)..))).)))--)))))))))..)..---........((((((((((.-----.-..((((.(((.(((((.(((.....))).)))))))))))).)))))-)))))--- ( -44.30, z-score = -2.64, R) >droAna3.scaffold_13248 2278028 147 + 4840945 -----UCUUAUACGGUAGUGUAGUCUUAAGUUCCAGGAGAGCGGAAAGUUG------GGUUUCUUUGAGCACAAGGAUAUU---AAGACGAGUUC--------CUAUGAGUCCAUU-----------UGUAGGCAGUUUUGCAGGGAAAUG--CGAUAAACUGCCAUCGAUUUAGGACUACU -----..............(((((((((((((...(((....((((.(((.------.((.((((((....)))))).)).---..)))...)))--------)......)))...-----------....((((((((((((......))--)))..)))))))...))))))))))))). ( -44.10, z-score = -2.13, R) >dp4.chrXL_group1e 8800775 152 - 12523060 ---------GCUUACAAAUGUUGUUUUAAGUUCCAAGAGAGCGGGAAGAGG-GGAUAUGGUUCUUCAAGCACUAGGAUAUU---AAAACCG--UUUC------CUUUGAGUCCAUUUUUAGU---G-GAUGCGCAGCUGGGA-AGGGGUUUUCCUUUCAGCUGGCAUUGAGGU-GGACU--- ---------......(((((..(((((((..(((...((.((..((((((.-........))))))..)).)).)))..))---)))))))--))).------.....((((((((((....---.-(((((.(((((((.(-((((....))))))))))))))))))))))-)))))--- ( -49.30, z-score = -1.74, R) >droPer1.super_17 1271651 152 - 1930428 ---------GCUUACAAAUGUUGUCUUAAGUUCCAAGAGAGCGGGAAGAGG-GGAUAUGUAUCUUCAAGCACUAGGAUAUU---AAAACCG--UUUC------CUUUGAGUCCAUUUUUAGU---G-GAUGCGCAGCUGGGA-AGGGGUUUUCCUUUCAGCUGGCAUUGAGGU-GGACU--- ---------.....((((((((.((((.......)))).))))(((((..(-(.....(((((((........))))))).---....)).--))))------)))))((((((((((....---.-(((((.(((((((.(-((((....))))))))))))))))))))))-)))))--- ( -49.60, z-score = -1.78, R) >droEre2.scaffold_4690 7386492 156 + 18748788 --------GUAUCUAUAGUGUUGUUUUAAUUUCCAGGAGAGCAGGAAGUGG----UUUAUAUCUUCAGGCACAAGGAUAGU--UAAAAAAAUUUCAC---UUAAUAUUAGUCCAUUUUU-----GGUAAUGCUCAGCACGACAGGGUAUUUAUAAACCGUCUGGCAUUGAGGA-GGACU--- --------........((((...(((((((((((..(...((..((((...----.......))))..)).)..))).)))--)))))......)))---).......(((((.(((((-----...(((((.(((.(((.................))))))))))))))))-)))))--- ( -30.93, z-score = 0.50, R) >droSec1.super_17 881374 148 + 1527944 --------UUGUCUUCAAUGUCGAAUUAAGUUCCAGGAGAGCGGGAAGUGG-----UUUUAUCUUCAAGCACAAGGAUAGU--UAAUACCUUUUC--------CUAUGAGUCCAUU-----AAAAGUUAUGCUCAGUUCGGC-UAAAAAUU-ACCAUCAACUGGCAUUGAGGA-GGACU--- --------((.(((((((((((((((...)))).((..((((((((((.((-----(.(((((((........))))))).--....))).))))--------)).(((((.....-----.........)))))))))..)-).......-..........)))))))))))-.))..--- ( -37.24, z-score = -1.06, R) >droWil1.scaffold_180702 1292995 170 - 4511350 -----CAAUAGUUUUCA-UGUGGUUUUAAGUUCCAAGAGAGCAGGAAGAGGAGGCUUAUAAUCUUCAGGCACUAGGAUAAUAUUAAAACCCUUUCAGAGGCUAUUCUCAGUCCAUUCUUUAAAAGAUUAUGCCCAGUU-GGU-UACAGAUUGAACCCUAACUGGCAUUGAGGU-GGACU--- -----.(((((((((..-.(.((((((((..(((...((.((..(((((..(......)..)))))..)).)).))).....))))))))).....)))))))))...(((((((.(((.........(((((.((((-(((-(........))))..))))))))).)))))-)))))--- ( -46.10, z-score = -1.05, R) >droVir3.scaffold_12970 9771061 154 + 11907090 -------GCAUAGCGUAAUGUGGUUUUAAGUUCCAAGAGAGCGGGAAGAGA----AUGAGAUCUUCAGGCACUAGGAUAUU---AAAACCCUAUAUU----UAUUAUGAGUCCAAUUGAAAU----GUAUGCUUAGCUCAGCAUGGAAAAC--AAGCUGGCUGGCAUUGAGGU-GGACU--- -------.(((((...(((((((((((((..(((...((.((..(((((..----......)))))..)).)).)))..))---))))))..)))))----..)))))((((((........----..(((((.(((.((((.((.....)--).)))))))))))).....)-)))))--- ( -47.76, z-score = -2.82, R) >droMoj3.scaffold_6473 5145330 157 - 16943266 AGAUUACAUUCAGAUAAAUGUGAUUUUAAGUUCCAAGAGAGCGGGAAGAGA-GUCUA---AUCUUCAAGCACUAGGAUAUU---AAAACC---UCAU------AUUUAAUAUCAGUCCAAUU---GAGAUGCGCAG-UGCAG-UGGAAAAUAAUACUUGCCUGGCAUUGAGGU-GGACU--- ..............(((((((((((((((..(((...((.((..(((((..-.....---.)))))..)).)).)))..))---))))..---))))------))))).....((((((...---..(((((.(((-.((((-((........))).)))))))))))....)-)))))--- ( -46.30, z-score = -3.27, R) >droGri2.scaffold_14853 7080432 157 - 10151454 ----UCAUGGCUGGAUAAUGUAGUUUUAAGUUCCAAGAGAGCAAGAAGAGA-AUGUG---AUCUUCAGGCACUAGGAUAUU---AAAACCUCAUUUA------AUAUAAGUCCAUUGUUAUUU--GAUAUGCUCAGCU-CAG-CAUGAAAUAUUUGCUGACCGGCAUUGAGGU-GGACU--- ----............((((..(((((((..(((...((.((..(((((..-.....---.)))))..)).)).)))..))---)))))..))))..------.....((((((((.......--...(((((..(.(-(((-((.........)))))).))))))...)))-)))))--- ( -40.02, z-score = -0.83, R) >consensus ________UUAUCUAUAAUGUUGUUUUAAGUUCCAAGAGAGCAGGAAGAGG____UUUAUAUCUUCAAGCACAAGGAUAUU___AAAACCAUUUCAC______AUAUGAGUCCAUUUUU______GUUAUGCUCAGCUCGGC_UGGAAAUUUUAAACCAACUGGCAUUGAGGU_GGACU___ ......................(((((.((((((....(.((..((((..............))))..)).)..))).)))...)))))...................(((((...............((((.(((........................))))))).......)))))... (-13.48 = -13.18 + -0.30)
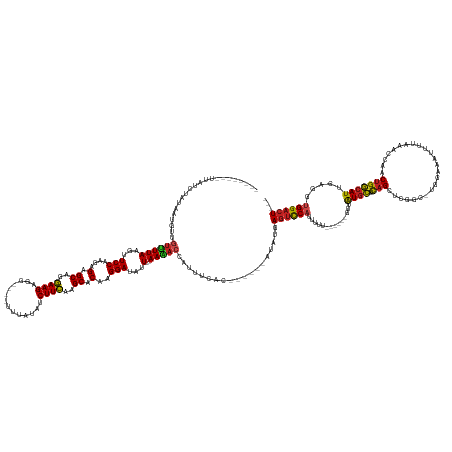
| Location | 17,039,753 – 17,039,911 |
|---|---|
| Length | 158 |
| Sequences | 12 |
| Columns | 182 |
| Reading direction | reverse |
| Mean pairwise identity | 65.24 |
| Shannon entropy | 0.71997 |
| G+C content | 0.39435 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -13.49 |
| Energy contribution | -13.29 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
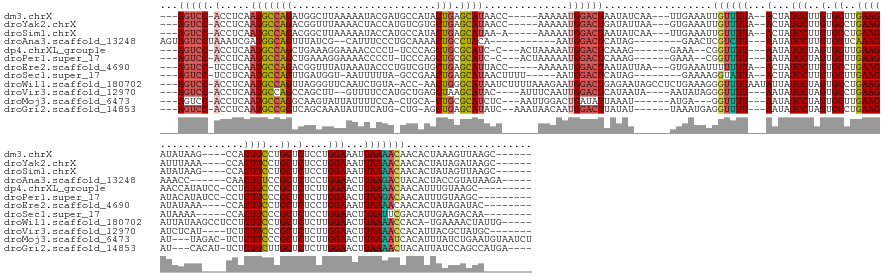
>dm3.chrX 17039753 158 - 22422827 ---AGUCC-ACCUCAAUGCCAGAUGGCUUAAAAAUACGAUGCCAUACUGAGCAUAACC-----AAAAAUGGACUAAUAUCAA---UUGAAAUUGUUUUA--ACUAUCCUUGUGCUUGAAGAUAUAAG----CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAAAGUUAAGC------ ---(((((-(.....((((((((((((.............))))).))).))))....-----.....))))))........---......((((((((--(...(((..(.((..((((.......----...))))..)).)....)))..)))))))))..............------ ( -34.06, z-score = -2.96, R) >droYak2.chrX 15649475 158 - 21770863 ---AGUCC-ACCUCAAUGCCAGACGGUUUAAAACUACCAUGUCGUGCUGAGCAUAACC-----AAAAAUGGACUGAUAUUAA---GUGAAAUUGUUUUA--ACUAUCCUUGUGCCUGAAGAUUUAAA----CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUAGAUAAGC------ ---(((((-(.....(((((((((((...............)))).))).))))....-----.....)))))).......(---(((...((((((((--(...(((..(.((..((((.......----...))))..)).)....)))..)))))))))))))..........------ ( -30.50, z-score = -1.29, R) >droSim1.chrX 13221011 157 - 17042790 ---AGUCC-ACCUCAAUGCCAGACGGCUUAAAAAUACCAUGCCAUACUGAGCAUAA-A-----AAAAAUGGACUAAUAUCAA---UUGAAAUUGUUUUA--ACUAUCCUUGUGCCUGAAGAUAUAAG----CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUAGUUAAGC------ ---(((((-(.....(((((((..(((.............)))...))).))))..-.-----.....))))))........---......((((((((--(...(((..(.((..((((.......----...))))..)).)....)))..)))))))))..............------ ( -29.64, z-score = -1.87, R) >droAna3.scaffold_13248 2278028 147 - 4840945 AGUAGUCCUAAAUCGAUGGCAGUUUAUCG--CAUUUCCCUGCAAAACUGCCUACA-----------AAUGGACUCAUAG--------GAACUCGUCUU---AAUAUCCUUGUGCUCAAAGAAACC------CAACUUUCCGCUCUCCUGGAACUUAAGACUACACUACCGUAUAAGA----- .(((((((((..((.((((((((((...(--((......))).))))))))....-----------.)).))....)))--------))....(((((---((..(((..(.((..((((.....------...))))..)).)....)))..)))))))....)))).........----- ( -33.50, z-score = -2.43, R) >dp4.chrXL_group1e 8800775 152 + 12523060 ---AGUCC-ACCUCAAUGCCAGCUGAAAGGAAAACCCCU-UCCCAGCUGCGCAUC-C---ACUAAAAAUGGACUCAAAG------GAAA--CGGUUUU---AAUAUCCUAGUGCUUGAAGAACCAUAUCC-CCUCUUCCCGCUCUCUUGGAACUUAAAACAACAUUUGUAAGC--------- ---(((((-(.....((((((((((.((((......)))-)..)))))).)))).-.---........))))))(((((------....--).(((((---((..(((.((.((..(((((.........-..)))))..))...)).)))..)))))))....)))).....--------- ( -39.76, z-score = -3.77, R) >droPer1.super_17 1271651 152 + 1930428 ---AGUCC-ACCUCAAUGCCAGCUGAAAGGAAAACCCCU-UCCCAGCUGCGCAUC-C---ACUAAAAAUGGACUCAAAG------GAAA--CGGUUUU---AAUAUCCUAGUGCUUGAAGAUACAUAUCC-CCUCUUCCCGCUCUCUUGGAACUUAAGACAACAUUUGUAAGC--------- ---(((((-(.....((((((((((.((((......)))-)..)))))).)))).-.---........))))))(((((------....--).(((((---((..(((.((.((..(((((.........-..)))))..))...)).)))..)))))))....)))).....--------- ( -39.66, z-score = -3.23, R) >droEre2.scaffold_4690 7386492 156 - 18748788 ---AGUCC-UCCUCAAUGCCAGACGGUUUAUAAAUACCCUGUCGUGCUGAGCAUUACC-----AAAAAUGGACUAAUAUUAA---GUGAAAUUUUUUUA--ACUAUCCUUGUGCCUGAAGAUAUAAA----CCACUUCCUGCUCUCCUGGAAAUUAAAACAACACUAUAGAUAC-------- ---(((((-.....((((((((((((...............)))).))).)))))...-----......))))).......(---(((...((.(((((--(...(((..(.((..((((.......----...))))..)).)....)))..)))))).))))))........-------- ( -24.70, z-score = -0.47, R) >droSec1.super_17 881374 148 - 1527944 ---AGUCC-UCCUCAAUGCCAGUUGAUGGU-AAUUUUUA-GCCGAACUGAGCAUAACUUUU-----AAUGGACUCAUAG--------GAAAAGGUAUUA--ACUAUCCUUGUGCUUGAAGAUAAAA-----CCACUUCCCGCUCUCCUGGAACUUAAUUCGACAUUGAAGACAA-------- ---.(((.-......(((((((((..((((-........-))))))))).))))....(((-----((((......(((--------((.((((.....--.....))))(.((..((((......-----...))))..)).))))))(((.....)))..))))))))))..-------- ( -30.20, z-score = -0.39, R) >droWil1.scaffold_180702 1292995 170 + 4511350 ---AGUCC-ACCUCAAUGCCAGUUAGGGUUCAAUCUGUA-ACC-AACUGGGCAUAAUCUUUUAAAGAAUGGACUGAGAAUAGCCUCUGAAAGGGUUUUAAUAUUAUCCUAGUGCCUGAAGAUUAUAAGCCUCCUCUUCCUGCUCUCUUGGAACUUAAAACCACA-UGAAAACUAUUG----- ---(((((-(.....((((((((...(((((.....).)-)))-.))).)))))..(((.....))).))))))...(((((..((......((((((((.....(((.((.((..(((((............)))))..))...)).)))..))))))))...-.))...))))).----- ( -37.50, z-score = -0.29, R) >droVir3.scaffold_12970 9771061 154 - 11907090 ---AGUCC-ACCUCAAUGCCAGCCAGCUU--GUUUUCCAUGCUGAGCUAAGCAUAC----AUUUCAAUUGGACUCAUAAUA----AAUAUAGGGUUUU---AAUAUCCUAGUGCCUGAAGAUCUCAU----UCUCUUCCCGCUCUCUUGGAACUUAAAACCACAUUACGCUAUGC------- ---(((((-(.....((((.(((((((.(--(.....)).)))).)))..))))..----........))))))((((...----.......((((((---((..(((.((.((..(((((......----..)))))..))...)).)))..)))))))).........)))).------- ( -36.17, z-score = -2.45, R) >droMoj3.scaffold_6473 5145330 157 + 16943266 ---AGUCC-ACCUCAAUGCCAGGCAAGUAUUAUUUUCCA-CUGCA-CUGCGCAUCUC---AAUUGGACUGAUAUUAAAU------AUGA---GGUUUU---AAUAUCCUAGUGCUUGAAGAU---UAGAC-UCUCUUCCCGCUCUCUUGGAACUUAAAAUCACAUUUAUCUGAAUGUAAUCU ---(((((-(.....((((((((((.((..........)-)))).-))).))))...---...))))))(((.......------....---((((((---((..(((.((.((..(((((.---.....-..)))))..))...)).)))..))))))))((((((....)))))).))). ( -37.50, z-score = -2.51, R) >droGri2.scaffold_14853 7080432 157 + 10151454 ---AGUCC-ACCUCAAUGCCGGUCAGCAAAUAUUUCAUG-CUG-AGCUGAGCAUAUC--AAAUAACAAUGGACUUAUAU------UAAAUGAGGUUUU---AAUAUCCUAGUGCCUGAAGAU---CACAU-UCUCUUCUUGCUCUCUUGGAACUUAAAACUACAUUAUCCAGCCAUGA---- ---(((((-(.....(((((((((((((.........))-)))-).))).))))...--.........)))))).....------..((((.((((((---((..(((.((.((..(((((.---.....-..)))))..))...)).)))..)))))))).))))............---- ( -38.63, z-score = -2.46, R) >consensus ___AGUCC_ACCUCAAUGCCAGACGGCUUAAAAUUUCCA_GCCGAGCUGAGCAUAAC______AAAAAUGGACUCAUAU______GUGAAAUGGUUUU___AAUAUCCUAGUGCCUGAAGAUAUAAA____CCACUUCCCGCUCUCCUGGAACUUAAAACAACAUUAAAGAGAA________ ...(((((.......(((((((........................))).))))...............)))))..................((((((.......(((..(.((..((((..............))))..)).)....)))....))))))..................... (-13.49 = -13.29 + -0.20)
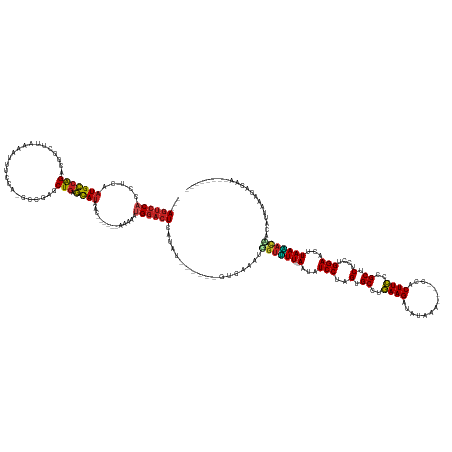
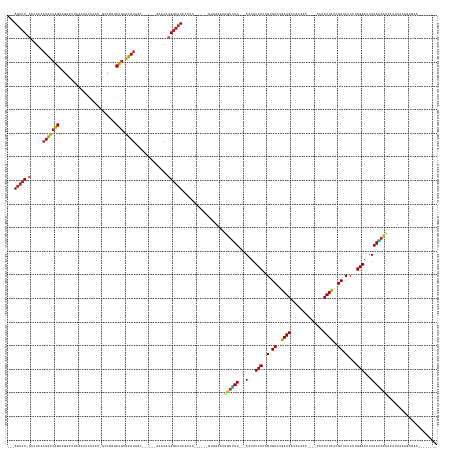
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:36 2011