| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,038,172 – 17,038,274 |
| Length | 102 |
| Max. P | 0.853790 |

| Location | 17,038,172 – 17,038,274 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Shannon entropy | 0.59791 |
| G+C content | 0.38865 |
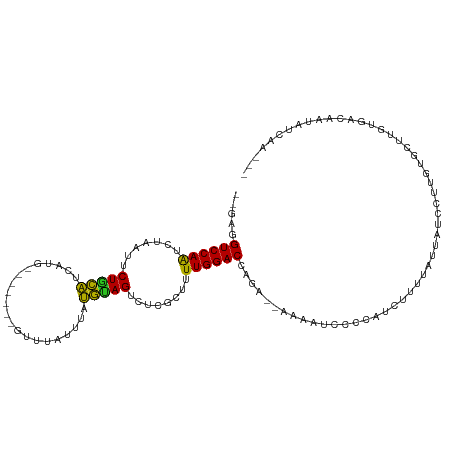
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.04 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
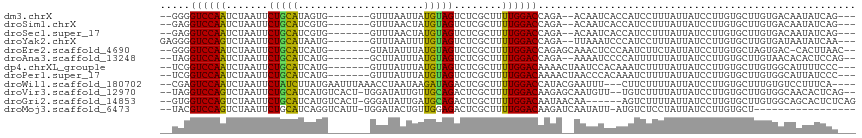
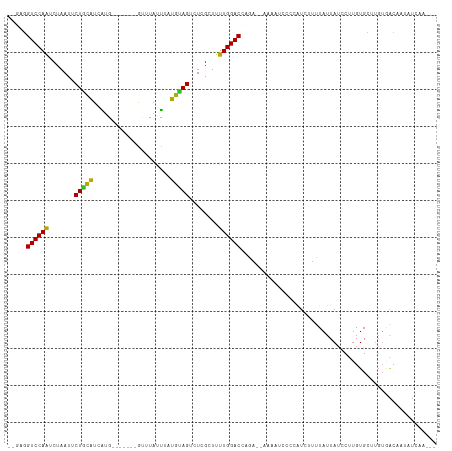
>dm3.chrX 17038172 102 - 22422827 --GGGGUCCAAUCUAAUUCUGCAUAGUG-------GUUUAAUUAUGUAGUCUCGCUUUUGGACCAGA--ACAAUCACCAUCCUUUAUUAUCCUUGUGCUUGUGACAAUAUCAG--- --..(((((((.......(((((((((.-------.....)))))))))........)))))))...--((((.(((.................))).))))...........--- ( -23.89, z-score = -1.62, R) >droSim1.chrX 13219410 102 - 17042790 --GAGGUCCAAUCUAAUUCUGCAUCGUG-------GUUUAACUAUGUAGUCUCGCUUUUGGACCAGA--ACAAUCACCAUCCUUUAUUAUCCUUGUGCUUGUGACAAUAUCAG--- --(((((((((.......((((((.((.-------.....)).))))))........)))))))...--((((.(((.................))).)))).......))..--- ( -21.99, z-score = -1.55, R) >droSec1.super_17 879945 102 - 1527944 --GAGGUCCAAUCUAAUUCUGCAUCGUG-------GUUUAACUAUGUAGUCUCGCUUUUGGACCAGA--ACAAUCACCAUCCUUUAUUAUCCUUGUGCUUGUGACAAUAUCAG--- --(((((((((.......((((((.((.-------.....)).))))))........)))))))...--((((.(((.................))).)))).......))..--- ( -21.99, z-score = -1.55, R) >droYak2.chrX 15648259 104 - 21770863 GAGGGGUCCAGUCUAAUUCUGCAUAAUG-------GUUUAAUUUUGUAGUCUCGCUUUUGGACCAGA--UUAAAUCCCAUCCUUUAUUAUCCUUGUGCUUGUGAUAAUAUCAA--- (((((((((((.......(((((.(((.-------.....))).)))))........))))))).((--(...)))....))))((((((((........).)))))))....--- ( -22.76, z-score = -1.25, R) >droEre2.scaffold_4690 7385244 104 - 18748788 --GGGGUCCAAUCUAAUUCUGCAUCAUG-------GUAUAUUUAUGUAGUCUCGCUUUUGGACCAGAGCAAACUCCCAAUCUUCUAUUAUCCUUGUGCUAGUGAC-CACUUAAC-- --(((((((((.......((((((.(..-------......).))))))........)))))))..((((.......(((.....))).......)))).....)-).......-- ( -19.40, z-score = 0.00, R) >droAna3.scaffold_13248 2276680 103 - 4840945 --UAGGUCCAAUCUAAUUCUGCAUCAUG-------GCUUAUUUAUGUAGUCUCGCUUUUGGACCAGA--AAAAUCCCCAUUUUUUAUUAUCCUUGUGCUUGUAACACACUCCAG-- --..(((((((.......((((((.(..-------......).))))))........))))))).((--(((((....)))))))........((((.......))))......-- ( -18.76, z-score = -1.59, R) >dp4.chrXL_group1e 8799343 104 + 12523060 --UCGGUCCAAUCUAAUUCUGCAUCAUG-------GUUUAUUUAUGUAGUCUCGCUUUUGGACAAAACUAAUCCACAAAUCUUUUAUUAUCCUUGUGCUUGUGGCAUUUUCCC--- --...((((((.......((((((.(..-------......).))))))........)))))).........((((((....................)))))).........--- ( -16.81, z-score = -1.03, R) >droPer1.super_17 1270230 104 + 1930428 --UCGGUCCAAUCUAAUUCUGCAUCAUG-------GUUUAUUUAUGUAGUCUCGCUUUUGGACAAAACUAACCCACAAAUCUUUUAUUAUCCUUGUGCUUGUGGCAUUAUCCC--- --...((((((.......((((((.(..-------......).))))))........)))))).........((((((....................)))))).........--- ( -17.01, z-score = -0.86, R) >droWil1.scaffold_180702 1291159 107 + 4511350 --CGAGUCCAAUCUAAUUCUAUCUUAUGAAUUUAAACCUAAUAAGAUAGACUCGCUUUUGGACCAUACGAAUUU---CUUCUUUUAUUAUCCUUGUGCUUUGUGUCCUUUCA---- --.(.((((((......((((((((((.............)))))))))).......)))))))(((((((...---(................)...))))))).......---- ( -18.83, z-score = -2.49, R) >droVir3.scaffold_12970 9769732 109 - 11907090 --UAGGUCCAGUCUAAUUCUGCAUCAUGUCACU-UGGAUAUUGUUGCAGACUCGCUUUUGGACAAGAGCAAUGUU--UGUCUUUUAUUAUCCUUGUGCUUGUGGCAACACUCAG-- --...((((((......((((((.((((((...-..)))).)).)))))).......))))))..(((((..(..--((........))..)...)))))(((....)))....-- ( -29.12, z-score = -1.43, R) >droGri2.scaffold_14853 7079175 107 + 10151454 --GUGGUCCAGUCUAAUUCUGCAUCAUGUCACU-GGGAUAUUGAUGCAGACUCGCUUUUGGACAAUAACAA------AGUCUUUUAUUAUCCUUGUGCUUGUGGCAGCACUCUCAG --((.((((((......(((((((((((((...-..)))).))))))))).......))))))....))..------.................(((((......)))))...... ( -30.02, z-score = -1.83, R) >droMoj3.scaffold_6473 5143951 95 + 16943266 --UACGUCCAGUCUAAUUCUGCAUCAGGUCAUU-UGGAUACUGUUGGAGACUCGCUUUUGGACAAGAUCAAUAUU-AUGUCUCCUAUUAUCCUUGUGCU----------------- --......(((.......))).....((.(((.-.(((((.....((((((......((((......))))....-..))))))...)))))..)))))----------------- ( -19.30, z-score = -0.10, R) >consensus __GAGGUCCAAUCUAAUUCUGCAUCAUG_______GUUUAUUUAUGUAGUCUCGCUUUUGGACCAGA__AAAAUCCCCAUCUUUUAUUAUCCUUGUGCUUGUGACAAUAUCAA___ .....((((((.......(((((.....................)))))........))))))..................................................... (-10.67 = -10.04 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:28 2011