| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,037,941 – 17,038,066 |
| Length | 125 |
| Max. P | 0.813542 |

| Location | 17,037,941 – 17,038,066 |
|---|---|
| Length | 125 |
| Sequences | 12 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Shannon entropy | 0.62843 |
| G+C content | 0.36921 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -12.39 |
| Energy contribution | -11.17 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17037941 125 - 22422827 GUAGUCUCGCUUUUGGACCAGAUUAAU--UUCCAUCCUUUAUUAUCCUUGUGC-UUGUAAUAUUG--CUUAGUUCAAUGCACAUGAGAAACGUAAAGGAACACUAGCUUUAAAUAAGUUAGGGACUUGUU .((((((.((.....).).))))))..--.(((.((((((((..(((.(((((-(((...((...--..))...))).))))).).))...))))))))...(((((((.....))))))))))...... ( -29.50, z-score = -1.70, R) >droSim1.chrX 13219179 125 - 17042790 GUAGUCUCGCUUUUGGACCAGAUUAAU--UCACAUCCUUUAUUAUCCUUGUGC-UUGUAAUAUUA--CUUAGUUCAAUGCACACGAGAAACGUAAAUGAACACUAGCUUUAAAUAAGUUAGGAACUUAUU ((..(((((.....(((...((.....--))...)))...........(((((-(((...((...--..))...))).)))))))))).))...........(((((((.....)))))))......... ( -22.20, z-score = -0.75, R) >droSec1.super_17 879714 125 - 1527944 GUAGUCUCGCUUUUGGACCAGAUUAAU--UCAAAUCCUUUAUUAUCCUUGUGC-UUGUAAUAUUA--CUUAGUUCAAUGCACACGCGAAACGUAAAUGAACACUAGCUUUAAAUAAGUUAGGAACUUAUU ...((.((((....(((...((.....--))...)))...........(((((-(((...((...--..))...))).))))).)))).))...........(((((((.....)))))))......... ( -23.30, z-score = -1.22, R) >droEre2.scaffold_4690 7385007 123 - 18748788 GUAGUCUCGCUUUUGGACCAGAACAAU--CACCAUCCUUUAUUAUCCUUGUGC-UUGUAAUAUCA--UUUAGUUCAAUGCACACGAGAAACGUAAAAGAACACUCG-AAUAAAUAAAUUAGCAACUUAU- ........(((...(((...((....)--)....)))((((((.....(((((-(((...((...--..))...))).)))))((((...............))))-))))))......))).......- ( -14.46, z-score = 0.80, R) >droYak2.chrX 15647851 124 - 21770863 GUAGUCUCGCUUUUGGACCAGAUUAAU--UCCCAUCCUUUAUUAUCCUUGUGC-UUGCAAUAUCA--UUCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAGCUUUAAAUAAGUUGAGAACUUGU- ....((((.((((((((...((.....--))...))).......(((.(((((-(((.(((....--....)))))).))))).).)).....)))))......(((((.....)))))))))......- ( -18.50, z-score = 0.62, R) >droAna3.scaffold_13248 2276628 119 - 4840945 GUAGUCUCGCUUUUGGACCAGAAAAAU--CCCCAUUUUUUAUUAUCCUUGUGC-UUGUAACACAC--UCCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAUCUUCUAA-AUCUUAAAAAU----- ((..(((((.....(((...(((((((--....)))))))....))).(((((-(((.(((....--....)))))).)))))))))).)).((.((((......)))).)).-...........----- ( -19.00, z-score = -1.66, R) >dp4.chrXL_group1e 8799095 123 + 12523060 GUAGUCUCGCUUUUGGACAAAACUAAUCCACAAAUCUUUUAUUAUCCUUGUGC-UUGUGGCACUAUCCCCUGUCCAAUGCACACGAGAAACGUAGAAGAACAUUAAAUAAGCAUCAGAAUAGAG------ ........((((.((((.........))))....((((((((..(((.(((((-(((.((((........))))))).))))).).))...)))))))).........))))............------ ( -24.30, z-score = -1.32, R) >droPer1.super_17 1269982 123 + 1930428 GUAGACUCGCUUUUGGACAAAACUAACCCACAAAUCUUUUAUUAUCCUUGUGC-UUGUGGCACUAUCCCCUGUCCAAUGCACACGAGAAACGUAGAAGAACAUUAAAUAAGCAUCAGAAUAGAG------ ........((((.(((...........)))....((((((((..(((.(((((-(((.((((........))))))).))))).).))...)))))))).........))))............------ ( -23.10, z-score = -1.15, R) >droWil1.scaffold_180702 1290924 115 + 4511350 AUAGACUCGCUUUUGGACUAGAUAAAU--UUUCUUCUUUUAUUAUCCUUGUGCUUUGUGUCCUUU--UAUCUGACAAGGCACACGAGAAACGUAAAUGAACAAAAGUACUAAACUUAUU----------- ........(((((((.....((.....--..))(((.(((((..(((.((((((((((.......--......)))))))))).).))...))))).))))))))))............----------- ( -21.62, z-score = -1.46, R) >droVir3.scaffold_12970 9769525 100 - 11907090 GCAGACUCGCUUUUGGACACGAGCAAUG--UAUGUCUUUUAUUAUCCUUGUGC-UUGUGGCAACA--CUCAGCUCAAUGCACACGAGAAACGUAGAAGAACAAUU------------------------- (((..((((..........))))...))--).((((((((((....((((((.-..(((....))--)...((.....)).))))))....))))))).)))...------------------------- ( -24.40, z-score = -0.74, R) >droMoj3.scaffold_6473 5143890 114 + 16943266 GGAGACUCGCUUUUGGACAAGAUCAAUA-UUAUGUCUCCUAUUAUCCUUGUGC-UUGUGGCAACA--CCCAGCUCAAUGCACACGAGAAACGUAGAAGAACAACUUGUUAAGCUAAUG------------ ((((((......((((......))))..-....)))))).......((((((.-..(((....))--)...((.....)).)))))).....(((...(((.....)))...)))...------------ ( -26.10, z-score = -1.04, R) >droGri2.scaffold_14853 7079136 104 + 10151454 GCAGACUCGCUUUUGGACAAUAACA------AAGUCUUUUAUUAUCCUUGUGC-UUGUGGCAGCACUCUCAGCUCAAUGCACACGAGAAACGUAGAAGAACAAAUCUUAUA------------------- ((......))(((((........))------)))((((((((..(((.(((((-(((.(((..........)))))).))))).).))...))))))))............------------------- ( -19.10, z-score = 0.38, R) >consensus GUAGUCUCGCUUUUGGACCAGAUUAAU__UCACAUCUUUUAUUAUCCUUGUGC_UUGUAACAUUA__CUCAGUUCAAUGCACACGAGAAACGUAAAAGAACACUAGCUUUAAAUAAGUUA_GAA______ ..................................((.(((((..(((.(((((.(((.(((..........)))))).))))).).))...))))).))............................... (-12.39 = -11.17 + -1.21)

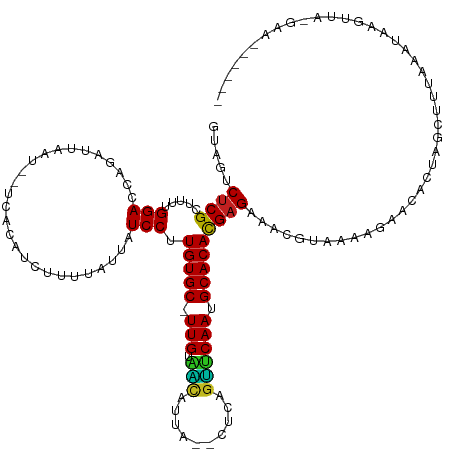
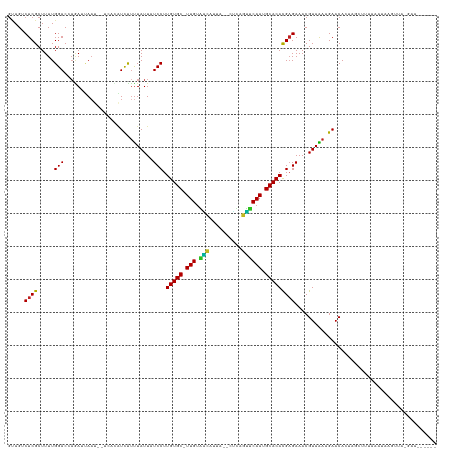
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:28 2011