| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,970,947 – 16,971,012 |
| Length | 65 |
| Max. P | 0.908631 |

| Location | 16,970,947 – 16,971,012 |
|---|---|
| Length | 65 |
| Sequences | 13 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
| Shannon entropy | 0.40957 |
| G+C content | 0.40031 |
| Mean single sequence MFE | -11.62 |
| Consensus MFE | -8.96 |
| Energy contribution | -9.12 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16970947 65 + 22422827 -------GAUAUAUUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGUUUUU- -------........(((((.(((....))).)).))).....(((.((((((......)))))).)))...- ( -11.90, z-score = -1.69, R) >apiMel3.GroupUn 73751267 66 - 399230636 -------GGUACCCUACCCAUCGCUGAUCCUUUGUGGUUUCCUCGCAUGCAUUACCAUCAAUGCCGGUGUCUG -------((((...))))...(((((......(((((.....))))).(((((......)))))))))).... ( -12.00, z-score = -0.01, R) >droSec1.super_17 804299 65 + 1527944 GAAAUAU-------UACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGUUUUU- ((((...-------.(((((.(((....))).)).))))))).(((.((((((......)))))).)))...- ( -12.90, z-score = -2.14, R) >droYak2.chrX 15582656 65 + 21770863 -------GAUAUAUUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGUUUUU- -------........(((((.(((....))).)).))).....(((.((((((......)))))).)))...- ( -11.90, z-score = -1.69, R) >droEre2.scaffold_4690 7324310 65 + 18748788 -------GAUAUAUUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGUUUUU- -------........(((((.(((....))).)).))).....(((.((((((......)))))).)))...- ( -11.90, z-score = -1.69, R) >droAna3.scaffold_13334 786673 68 - 1562580 CAAAUAUC----CAUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGAUUUU- ........----...(((((.(((....))).)).))).........((((((......)))))).......- ( -10.70, z-score = -1.34, R) >droPer1.super_30 878695 65 - 958394 --GAAAUC-----GUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAAUUUU- --((((((-----(((.....(((....))).)))))))))......((((((......)))))).......- ( -11.70, z-score = -1.51, R) >dp4.chrXL_group1e 1116193 65 + 12523060 --GAAAUC-----GUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAAUUUU- --((((((-----(((.....(((....))).)))))))))......((((((......)))))).......- ( -11.70, z-score = -1.51, R) >droWil1.scaffold_181096 8470301 70 - 12416693 --AAAAUCUUUUCGUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAAUGUU- --.............(((((.(((....))).)).))).....((((((((((......))))))...))))- ( -10.80, z-score = -1.11, R) >droVir3.scaffold_12970 9698605 69 + 11907090 ---AAAUUGUAAUUUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAGUUUU- ---....(((.....(((((.(((....))).)).)))......)))((((((......)))))).......- ( -12.00, z-score = -1.39, R) >droMoj3.scaffold_6473 5073104 69 - 16943266 ---AAAUUGUAAUUUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAGUUUU- ---....(((.....(((((.(((....))).)).)))......)))((((((......)))))).......- ( -12.00, z-score = -1.39, R) >droGri2.scaffold_14853 9810495 69 - 10151454 ---AAUUUGUAAUUUACCCAUCGUUCAUUCGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAAGUUUU- ---....(((.....(((((.((......)).)).)))......)))((((((......)))))).......- ( -10.70, z-score = -1.10, R) >triCas2.ChLG3 6380297 58 + 32080666 --------------UACCCAUCGCUCAUCCGUUGAGGUUUCCUUGCAUGCAUUACCAUCAAUGCAAGUCUCU- --------------.........((((.....)))).....(((((((............))))))).....- ( -10.90, z-score = -1.74, R) >consensus ___AAAUC_UA__UUACCCAUCGUUCAUACGCUGCGGUUUUCUAGCAUGCAUUACCAUCAAUGCAAGUUUUU_ ...............(((((.((......)).)).))).........((((((......))))))........ ( -8.96 = -9.12 + 0.15)

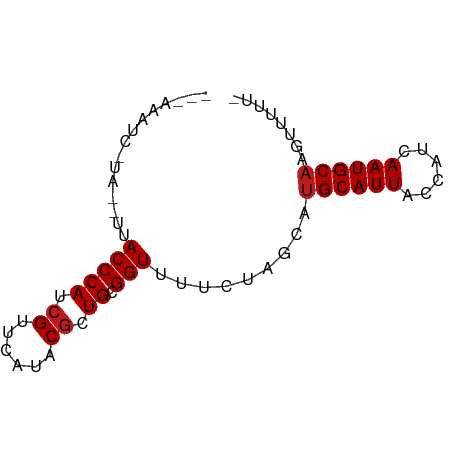
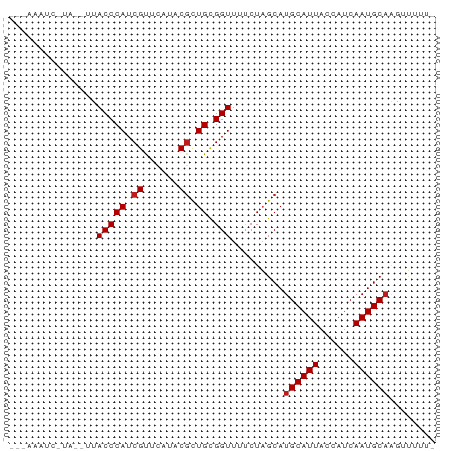
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:20 2011