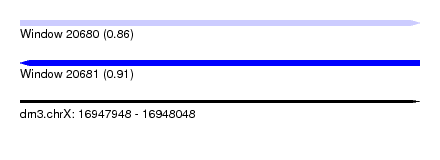
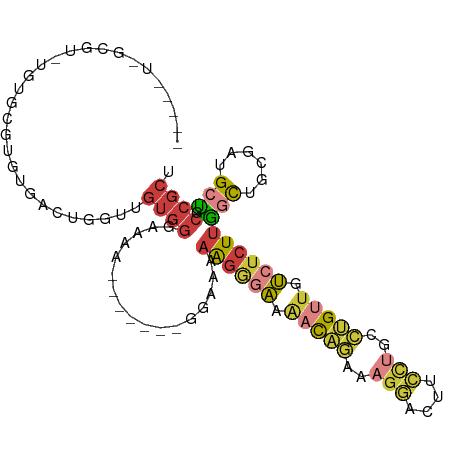
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,947,948 – 16,948,048 |
| Length | 100 |
| Max. P | 0.912556 |

| Location | 16,947,948 – 16,948,048 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 58.88 |
| Shannon entropy | 0.80595 |
| G+C content | 0.51868 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -5.82 |
| Energy contribution | -8.13 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857431 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 16947948 100 + 22422827 AGCGGCAGCAUCGCAGCCAAGAGACAACAGGCAGGAAGUCCUUUCUGUUUUCCCUUUUUCC------AUUUUCCCACAACCAGACACACGCACACACGCAACGCUA ((((((.((...)).)))(((.((.((((((.(((....))).)))))).)).))).....------......................((......))...))). ( -21.80, z-score = -2.08, R) >droSim1.chrX 13126059 101 + 17042790 AGCGGCAGCAUCGCAGCCAAGAGACAACAGGCAGGAAGUCCUUUCUGUUUUCCCUUUUUCCAU-----UUUCCCCACAACCAGUCACACGCACACACGCAACGCUA ((((((.((...)).)))(((.((.((((((.(((....))).)))))).)).))).......-----.....................((......))...))). ( -21.80, z-score = -1.97, R) >droSec1.super_17 782073 86 + 1527944 AGCGGCAGCAUCGCAGUCAAGAGACAACAGGCAGGAAGUCCUUUCUGUUUUCCCUUUUUC------------UCCACAACCAGUCACACGCA---ACGCUA----- ((((...((......(((....)))((((((.(((....))).))))))...........------------.................)).---.)))).----- ( -19.00, z-score = -1.15, R) >droYak2.chrX 15559758 106 + 21770863 AGCGGCAGCCUCGCAGCCAAGAGACAACAGGCCGGAAGUCCCUUCUGUUUUCCCUUUUCCCUUUGCCAUUUUCCCACAACCAAACACUCGCACACACGCAAUGCUA (((((((((......)).(((.((.((((((..((....))..)))))).)).))).......))))......................((......))...))). ( -21.00, z-score = -0.47, R) >droEre2.scaffold_4690 7301948 87 + 18748788 AGCGGCAGCAUCGCAGCCAAGAGACAACAGGCAGGAAGUCCUUUCUGUUUUCCCUUUUCC-------AUUUUCCCACAGCCAAACACAAGGCUA------------ ...(((.((...)).)))(((.((.((((((.(((....))).)))))).)).)))....-------..........((((........)))).------------ ( -22.80, z-score = -2.09, R) >droAna3.scaffold_13334 761720 75 - 1562580 AGCGGCGGCUGGGAAACCGAGAGACAACACAUA-----UCCUGUCAGGCUUUUUUAUGUCC-------UUACACCACAUCCA-CCGCA------------------ .((((.((..((....))(((.((((.......-----.((.....))........)))))-------)).........)).-)))).------------------ ( -21.19, z-score = -1.98, R) >droWil1.scaffold_181096 6190848 92 + 12416693 ------CGCAACACAAUAAAGUGCCAACCAGCCACGCCCACUCAUAAAGUGUGCCUCCCAUUUACACGGCAACAUAAAGUGAGUAACGCGCGUGGCAA-------- ------.....(((......))).......((((((((.((((((...(((((((............))).))))...))))))...).)))))))..-------- ( -28.30, z-score = -3.04, R) >consensus AGCGGCAGCAUCGCAGCCAAGAGACAACAGGCAGGAAGUCCUUUCUGUUUUCCCUUUUUCC_______UUUUCCCACAACCAGACACACGCACA_ACGC_A_____ ...(((.((...)).)))(((.((.((((((.(((....))).)))))).)).))).................................................. ( -5.82 = -8.13 + 2.31)

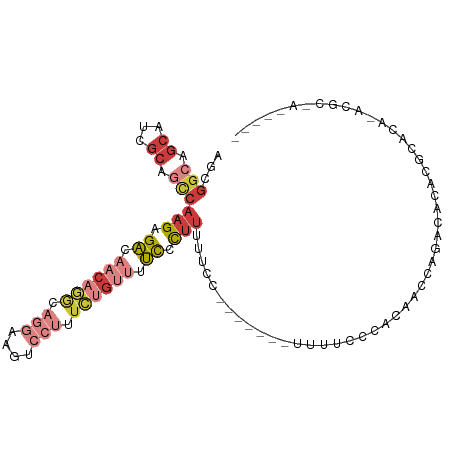
| Location | 16,947,948 – 16,948,048 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 58.88 |
| Shannon entropy | 0.80595 |
| G+C content | 0.51868 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.76 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912556 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 16947948 100 - 22422827 UAGCGUUGCGUGUGUGCGUGUGUCUGGUUGUGGGAAAAU------GGAAAAAGGGAAAACAGAAAGGACUUCCUGCCUGUUGUCUCUUGGCUGCGAUGCUGCCGCU ((((((((((.((...(((...(((......)))...))------)....((((((.(((((..(((....)))..))))).)))))).))))))))))))..... ( -31.80, z-score = -1.57, R) >droSim1.chrX 13126059 101 - 17042790 UAGCGUUGCGUGUGUGCGUGUGACUGGUUGUGGGGAAA-----AUGGAAAAAGGGAAAACAGAAAGGACUUCCUGCCUGUUGUCUCUUGGCUGCGAUGCUGCCGCU (((((((((.......(.((..(....)..)).)....-----..((...((((((.(((((..(((....)))..))))).))))))..)))))))))))..... ( -30.50, z-score = -0.87, R) >droSec1.super_17 782073 86 - 1527944 -----UAGCGU---UGCGUGUGACUGGUUGUGGA------------GAAAAAGGGAAAACAGAAAGGACUUCCUGCCUGUUGUCUCUUGACUGCGAUGCUGCCGCU -----((((((---((((.((.((.....))...------------....((((((.(((((..(((....)))..))))).)))))).))))))))))))..... ( -28.70, z-score = -2.35, R) >droYak2.chrX 15559758 106 - 21770863 UAGCAUUGCGUGUGUGCGAGUGUUUGGUUGUGGGAAAAUGGCAAAGGGAAAAGGGAAAACAGAAGGGACUUCCGGCCUGUUGUCUCUUGGCUGCGAGGCUGCCGCU .((((((.(((....))))))))).....((((.(...(.(((....(..((((((.(((((...((....))...))))).))))))..)))).)...).)))). ( -27.80, z-score = 1.58, R) >droEre2.scaffold_4690 7301948 87 - 18748788 ------------UAGCCUUGUGUUUGGCUGUGGGAAAAU-------GGAAAAGGGAAAACAGAAAGGACUUCCUGCCUGUUGUCUCUUGGCUGCGAUGCUGCCGCU ------------.((((........))))((((......-------....((((((.(((((..(((....)))..))))).))))))(((......))).)))). ( -29.60, z-score = -2.11, R) >droAna3.scaffold_13334 761720 75 + 1562580 ------------------UGCGG-UGGAUGUGGUGUAA-------GGACAUAAAAAAGCCUGACAGGA-----UAUGUGUUGUCUCUCGGUUUCCCAGCCGCCGCU ------------------.((((-(((.((.(((((..-------..))).....(((((.(((((.(-----....).)))))....))))))))).))))))). ( -24.30, z-score = -1.42, R) >droWil1.scaffold_181096 6190848 92 - 12416693 --------UUGCCACGCGCGUUACUCACUUUAUGUUGCCGUGUAAAUGGGAGGCACACUUUAUGAGUGGGCGUGGCUGGUUGGCACUUUAUUGUGUUGCG------ --------..(((((((.(...(((((.....(((.(((............)))))).....)))))).))))))).....(((((......)))))...------ ( -30.00, z-score = -0.68, R) >consensus _____U_GCGU_UGUGCGUGUGACUGGUUGUGGGAAAA_______GGAAAAAGGGAAAACAGAAAGGACUUCCUGCCUGUUGUCUCUUGGCUGCGAUGCUGCCGCU .............................((((.................((((((.(((((..(((....)))..))))).))))))(((......))).)))). (-11.85 = -12.76 + 0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:16 2011