| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,903,667 – 16,903,816 |
| Length | 149 |
| Max. P | 0.934131 |

| Location | 16,903,667 – 16,903,776 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Shannon entropy | 0.06418 |
| G+C content | 0.39745 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -22.20 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16903667 109 - 22422827 ACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCAUUGUUUUUUGGUAUCUGCGCUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ...........((((.((.((((((...((...(((((......((((((....(((((..((....))..)))))..))))))))))).))...)))))).)).)))) ( -24.40, z-score = -1.69, R) >droEre2.scaffold_4690 7258895 108 - 18748788 ACUUCAUCUCCCCUGGGAUAUUUGACUUGUUCUGUUUUCCCUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ............(((.((.((((((...((....(((((....((((-(.....(((((..((....))..)))))))))).)))))...))...)))))).)).))). ( -24.10, z-score = -1.27, R) >droYak2.chrX 15513448 108 - 21770863 ACUUCAUCUCCCCUGGGAUAUUUGAGUUGUUCUGUUUUUCCUCUCUG-UUUUCGGUAUCUGCACUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ............(((.((.((((((...((...(((((((...((((-(.....(((((..((....))..)))))))))).))))))).))...)))))).)).))). ( -28.40, z-score = -2.62, R) >droSec1.super_17 737365 108 - 1527944 ACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ...........((((.((.((((((...((....(((((....((((-(.....(((((..((....))..)))))))))).)))))...))...)))))).)).)))) ( -24.70, z-score = -1.82, R) >droSim1.chrX 13085611 108 - 17042790 ACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ...........((((.((.((((((...((....(((((....((((-(.....(((((..((....))..)))))))))).)))))...))...)))))).)).)))) ( -24.70, z-score = -1.82, R) >consensus ACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG_UUUUCGGUAUCUGCGCUGUUGUGGGUACAUAGACGAAAAAUUGCUUAUCAAAUUUCGCAGA ............(((.((.((((((...((....(((((....((((.......(((((..((....))..))))).)))).)))))...))...)))))).)).))). (-22.20 = -21.92 + -0.28)

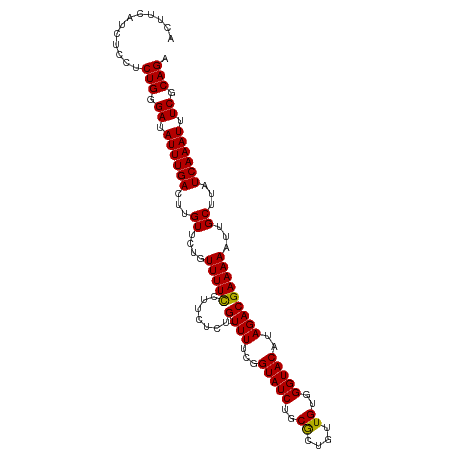
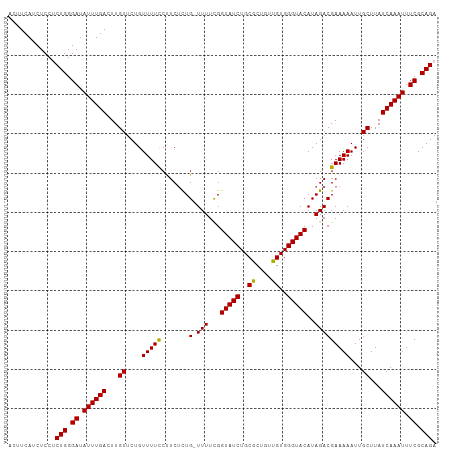
| Location | 16,903,697 – 16,903,816 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Shannon entropy | 0.39751 |
| G+C content | 0.39915 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -14.10 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16903697 119 + 22422827 UAUGUACCCACAACAGCGCAGAUACCAAAAAACAAUGAAGGAAAACAGAACAAGUCAAAUAUCCCAGAGGAGAUGAAGUUCUGUUGCCAGGAAUACAAAAAUUUAUGUACCGGAAAAUG ........................((.............((..((((((((...(((....(((....)))..))).)))))))).)).((..((((........))))))))...... ( -20.00, z-score = -1.35, R) >droAna3.scaffold_13334 709272 91 - 1562580 -----------------------GCUGAAAG---AAGAUAGAGAGGAGCACAAGUCAAAUAUCCCAAGGGAUAUCAAGAGUUGUGCCAUUAAAUGUACUAGG--AUAUGCCAGGAUAUG -----------------------........---...........(.((((((.((..((((((....))))))...)).))))))).....((((.((.((--.....)).)))))). ( -20.80, z-score = -1.88, R) >droEre2.scaffold_4690 7258925 118 + 18748788 UAUGUACCCACAACAGCGCAGAUACCGAAAA-CAGAGAGGGAAAACAGAACAAGUCAAAUAUCCCAGGGGAGAUGAAGUUCUGUUGCCAGGAAUACAAAAAUUUGUGUACCGAAAUAUG ...............((((((((.((.....-......))(..((((((((...(((....(((....)))..))).))))))))..)............))))))))........... ( -24.20, z-score = -1.33, R) >droYak2.chrX 15513478 118 + 21770863 UAUGUACCCACAACAGUGCAGAUACCGAAAA-CAGAGAGGAAAAACAGAACAACUCAAAUAUCCCAGGGGAGAUGAAGUUCUGUUCCCAGAAAUACACAAAUUUAUGUACCGGAAAAUG ..(((((........)))))....(((....-....(.(((...(((((((...(((....(((....)))..))).))))))))))).....((((........)))).)))...... ( -23.90, z-score = -1.83, R) >droSec1.super_17 737395 118 + 1527944 UAUGUACCCACAACAGCGCAGAUACCGAAAA-CAGAGAAGGAAAACAGAACAAGUCAAAUAUCCCAGAGGAGAUGAAGUUCUGUUGCCAGGAAUACAAAAAUUUAUGUACCGGAAAAUG ........................(((....-.......((..((((((((...(((....(((....)))..))).)))))))).)).....((((........)))).)))...... ( -20.20, z-score = -1.41, R) >droSim1.chrX 13085641 118 + 17042790 UAUGUACCCACAACAGCGCAGAUACCGAAAA-CAGAGAAGGAAAACAGAACAAGUCAAAUAUCCCAGAGGAGAUGAAGUUCUGUUGCCAGGAAUACAAAAAUUUAUGUACCGGAAAAUG ........................(((....-.......((..((((((((...(((....(((....)))..))).)))))))).)).....((((........)))).)))...... ( -20.20, z-score = -1.41, R) >consensus UAUGUACCCACAACAGCGCAGAUACCGAAAA_CAGAGAAGGAAAACAGAACAAGUCAAAUAUCCCAGAGGAGAUGAAGUUCUGUUGCCAGGAAUACAAAAAUUUAUGUACCGGAAAAUG ........................(((............((...(((((((...(((....(((....)))..))).)))))))..)).....((((........)))).)))...... (-14.10 = -13.88 + -0.22)

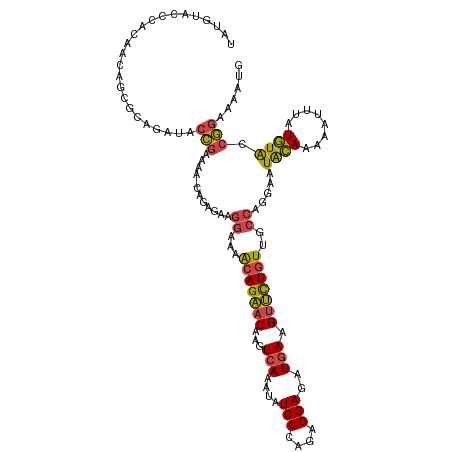
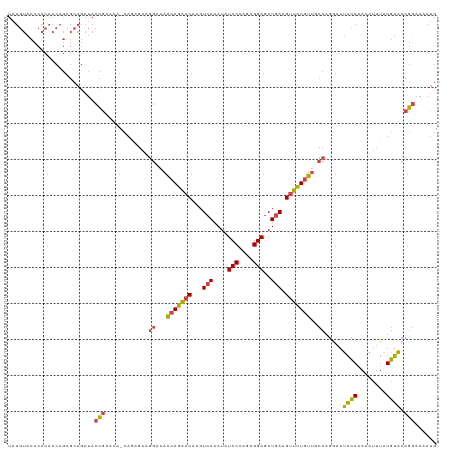
| Location | 16,903,697 – 16,903,816 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Shannon entropy | 0.39751 |
| G+C content | 0.39915 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.19 |
| Energy contribution | -18.85 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
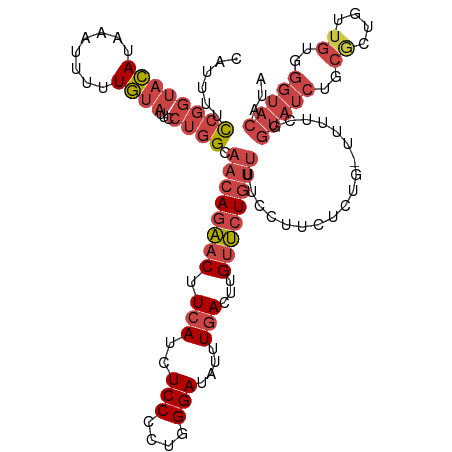
>dm3.chrX 16903697 119 - 22422827 CAUUUUCCGGUACAUAAAUUUUUGUAUUCCUGGCAACAGAACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCAUUGUUUUUUGGUAUCUGCGCUGUUGUGGGUACAUA ......((((((((........))))...)))).((((((((.(((..(((....)))....)))...))))))))..................(((((..((....))..)))))... ( -25.90, z-score = -1.64, R) >droAna3.scaffold_13334 709272 91 + 1562580 CAUAUCCUGGCAUAU--CCUAGUACAUUUAAUGGCACAACUCUUGAUAUCCCUUGGGAUAUUUGACUUGUGCUCCUCUCUAUCUU---CUUUCAGC----------------------- ......((((.....--.))))..........(((((((.((..(((((((....))))))).)).)))))))............---........----------------------- ( -20.50, z-score = -2.50, R) >droEre2.scaffold_4690 7258925 118 - 18748788 CAUAUUUCGGUACACAAAUUUUUGUAUUCCUGGCAACAGAACUUCAUCUCCCCUGGGAUAUUUGACUUGUUCUGUUUUCCCUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUA ........((...((((....))))...)).((.((((((((.(((..(((....)))....)))...))))))))..)).......-......(((((..((....))..)))))... ( -26.20, z-score = -1.19, R) >droYak2.chrX 15513478 118 - 21770863 CAUUUUCCGGUACAUAAAUUUGUGUAUUUCUGGGAACAGAACUUCAUCUCCCCUGGGAUAUUUGAGUUGUUCUGUUUUUCCUCUCUG-UUUUCGGUAUCUGCACUGUUGUGGGUACAUA ......((((((((((....))))))...))))(((((((((((((..(((....)))....))))..)))))))))..........-......(((((..((....))..)))))... ( -31.60, z-score = -2.43, R) >droSec1.super_17 737395 118 - 1527944 CAUUUUCCGGUACAUAAAUUUUUGUAUUCCUGGCAACAGAACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUA .......(((((((........)))))....((.((((((((.(((..(((....)))....)))...))))))))..)).......-....))(((((..((....))..)))))... ( -26.10, z-score = -1.78, R) >droSim1.chrX 13085641 118 - 17042790 CAUUUUCCGGUACAUAAAUUUUUGUAUUCCUGGCAACAGAACUUCAUCUCCUCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG-UUUUCGGUAUCUGCGCUGUUGUGGGUACAUA .......(((((((........)))))....((.((((((((.(((..(((....)))....)))...))))))))..)).......-....))(((((..((....))..)))))... ( -26.10, z-score = -1.78, R) >consensus CAUUUUCCGGUACAUAAAUUUUUGUAUUCCUGGCAACAGAACUUCAUCUCCCCUGGGAUAUUUGACUUGUUCUGUUUUCCUUCUCUG_UUUUCGGUAUCUGCGCUGUUGUGGGUACAUA ......((((((((........))))...)))).((((((((.(((..(((....)))....)))...))))))))..................(((((((((....)))))))))... (-17.19 = -18.85 + 1.66)

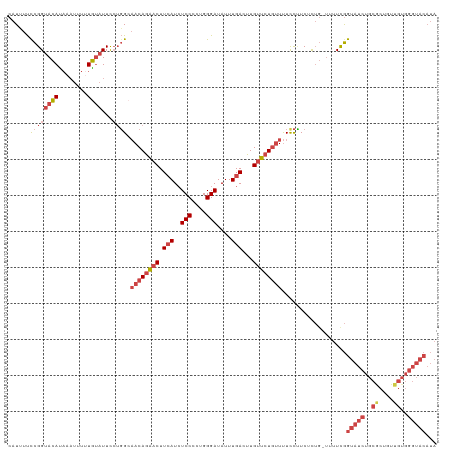
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:07 2011