| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,620,811 – 11,620,909 |
| Length | 98 |
| Max. P | 0.787611 |

| Location | 11,620,811 – 11,620,909 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.00 |
| Shannon entropy | 0.64222 |
| G+C content | 0.33085 |
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -7.00 |
| Energy contribution | -6.61 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787611 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
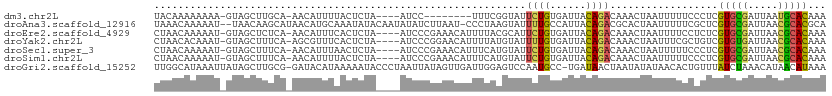
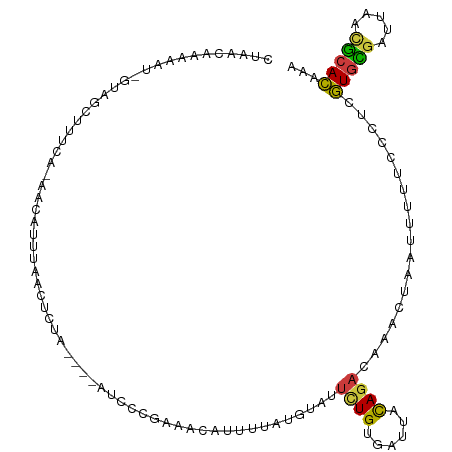
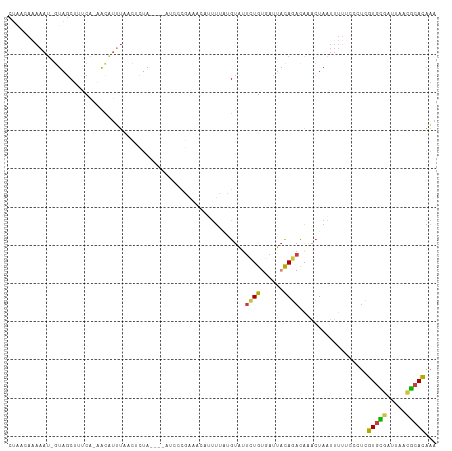
>dm3.chr2L 11620811 98 - 23011544 UACAAAAAAAA-GUAGCUUGCA-AACAUUUUACUCUA----AUCC--------UUUCGGUAUUCUGUGAUUACAGACAAACUAAUUUUUCCCUCGUGCGAUUAAUGCACAAA ..........(-((((..((..-..))..)))))...----....--------....(((..(((((....)))))...)))............(((((.....)))))... ( -12.10, z-score = -0.27, R) >droAna3.scaffold_12916 9338405 109 + 16180835 UAAACAAAAAU--UAACAAGCAUAACAUGCAAAUAUACAAUAUAUCUUAAU-CCCUAAGUAUUUUGCCAUUACAGACGCACUAAUUUUUCGCUCGUGCGAUUAACGCACGCA .....((((((--((....((((...)))).......(((.(((.((((..-...))))))).)))...............))))))))....((((((.....)))))).. ( -15.80, z-score = -1.19, R) >droEre2.scaffold_4929 12851028 106 + 26641161 CUAACAAAAAU-GUAGCUCUCA-AACAUUUCACUCUA----AUCCCGAAACAUUUUACGCAUUCUGUGAUUACAGACAAACUAAUUUUCCUCUCGUGCGAUUAACGCACAAA ......(((((-((..(.....-..............----.....)..)))))))......(((((....)))))..................(((((.....)))))... ( -13.06, z-score = -1.43, R) >droYak2.chr2L 8043350 106 - 22324452 CUAACACAAAU-GUAGCUUUCA-AGCGUUUCACUCUA----AUCCCGGAACAUUUUAUGUAUUUUGUGAUUACAGACAAACUAAUUUCGCUGUCGUGUGAUUAACGCACAAA ....((((((.-...(((....-)))(((((......----.....)))))...........))))))......((((............))))(((((.....)))))... ( -18.50, z-score = -0.47, R) >droSec1.super_3 7004908 106 - 7220098 CUAACAAAAAU-GUAGCUUUCA-AACAUUUAACUCUA----AUCCCGAAACAUUUCAUGUAUUCUGUGAUUACAGACAAACUAAUUUUUCCCUCGUGCGAUUAACGCACAAA .......((((-((........-.))))))..((.((----((((.(((((((...)))).))).).))))).))...................(((((.....)))))... ( -15.80, z-score = -2.32, R) >droSim1.chr2L 11428338 106 - 22036055 CUAACAAAAAU-GUAGCUUUCA-AACAUUUUACUCUA----AUCCCGAAACAUUUCAUGUAUUCUGUGAUUACAGACAAACUAAUUUUUCCCUCGUGCGAUUAACGCACAAA ......(((((-((........-.))))))).((.((----((((.(((((((...)))).))).).))))).))...................(((((.....)))))... ( -16.70, z-score = -2.63, R) >droGri2.scaffold_15252 9445422 110 - 17193109 UUGGCAUAAAUUAUAGCUUGCG-GAUACAUAAAAAUACCCUAAUUAUAGUUGAUUGGAGUCCAAUGCC-UGAUAACUAAUAUAUAACACUGUUUAUCUAAACAUAACAUAAA ..(((((........(....)(-(((..((....))...(((((((....))))))).)))).)))))-....................(((((....)))))......... ( -12.70, z-score = 0.90, R) >consensus CUAACAAAAAU_GUAGCUUUCA_AACAUUUAACUCUA____AUCCCGAAACAUUUUAUGUAUUCUGUGAUUACAGACAAACUAAUUUUUCCCUCGUGCGAUUAACGCACAAA ..............................................................((((......))))..................(((((.....)))))... ( -7.00 = -6.61 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:08 2011