
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,901,702 – 16,901,786 |
| Length | 84 |
| Max. P | 0.500000 |

| Location | 16,901,702 – 16,901,786 |
|---|---|
| Length | 84 |
| Sequences | 9 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Shannon entropy | 0.45839 |
| G+C content | 0.48640 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.56 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 16901702 84 + 22422827 UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU--CAUGAGGGCCCAUUGCCAUUG---CGUCUAAGCCAUUGUCUUU-----GGGUCCUG .((((((....))))))...(((.(((.....))))--))..((((((((..(.((..(---(......))...)).)..)-----))))))). ( -28.70, z-score = -2.25, R) >droSim1.chrX 13083646 84 + 17042790 UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU--CAUGAGGGCCCAUUGCCAUUG---CGUCUAAGCCAUUGUCUUU-----GGGUCCUG .((((((....))))))...(((.(((.....))))--))..((((((((..(.((..(---(......))...)).)..)-----))))))). ( -28.70, z-score = -2.25, R) >droSec1.super_17 734098 84 + 1527944 UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU--CAUGAGGGCCCAUUGCCGUUG---CGUCUAAGCCAUUGUCUUU-----GGGUCCUG .((((((....))))))...(((.(((.....))))--))..((((((((..((.((.(---(......)).)).).)..)-----))))))). ( -28.10, z-score = -1.93, R) >droYak2.chrX 15511438 79 + 21770863 UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU--CAUGAGGGCCCAUUGC--------CGUCUAAGCCAUUGUCUUU-----GGGUCCUG .((((((....))))))(((..((((....((((((--...((.(((.....))--------).))..)))))).).))).-----.))).... ( -29.10, z-score = -3.03, R) >droEre2.scaffold_4690 7257037 79 + 18748788 UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU--CAUGAGGGCCCGUUGC--------CGUCUAAGCCAUUGUCUUU-----GGGUCCUG .((((((....))))))(((..((((....((((((--...((.(((.....))--------).))..)))))).).))).-----.))).... ( -29.10, z-score = -2.90, R) >droAna3.scaffold_13334 700343 92 - 1562580 UGCGUGAAAAUUUACGCGCUUGACGCUUUUAUGGCU--CAUGAGGGCCCAUUGCCGUUGCGUCGUCUAAGCCAUUGUCUUUGGCUUUGGUCCUG .((((((....))))))....(((((....(((((.--.(((......))).))))).)))))(.((((((((.......)))).)))).)... ( -30.60, z-score = -1.92, R) >droWil1.scaffold_181096 6136814 80 + 12416693 GGAGAAAAAAUGUACGCGCUUGACGCUUUUAUGGCUUUCAUGGGGGUCCAUUGUCUACUCUACUUUUGGUCUGCAUUUUA-------------- ......((((((((.(((.....)))......((((......(((((.........)))))......)))))))))))).-------------- ( -14.70, z-score = 0.31, R) >droPer1.super_30 797529 87 - 958394 UGCGUGAAAAUUUACGCGCUCGACGCUUUUAUGGCU--CAUGAGAGGACCUAAGCCAUA---GCCCUAUCCCAUUGUCUUUUGC--AGGUUCUG .((((((....)))))).((((..(((.....))).--..))))((((((((((.((..---............)).)))....--))))))). ( -19.04, z-score = 0.15, R) >dp4.chrXL_group1e 1034528 87 + 12523060 UGCGUGAAAAUUUACGCGCUCGACGCUUUUAUGGCU--CAUGAGAGGACCUAAGCCAUA---GCCCUAUCCCAUUGUCUUUUGC--AGGUUCUG .((((((....)))))).((((..(((.....))).--..))))((((((((((.((..---............)).)))....--))))))). ( -19.04, z-score = 0.15, R) >consensus UGCGUGAAAAUUUACGCGCUUGAAGCUUUUAUGGCU__CAUGAGGGCCCAUUGCC_UUG___CGUCUAAGCCAUUGUCUUU_____GGGUCCUG .((((((....)))))).......(((((((((.....)))))))))............................................... (-12.44 = -12.56 + 0.11)

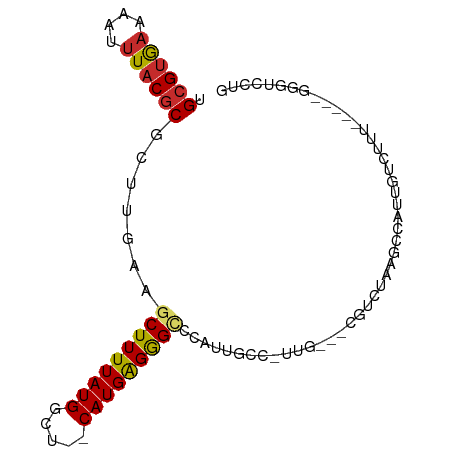
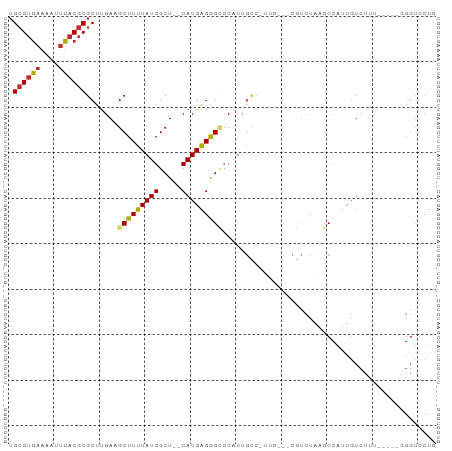
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:04 2011