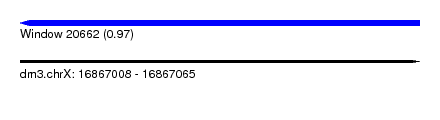
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,867,008 – 16,867,065 |
| Length | 57 |
| Max. P | 0.974120 |

| Location | 16,867,008 – 16,867,065 |
|---|---|
| Length | 57 |
| Sequences | 13 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Shannon entropy | 0.27324 |
| G+C content | 0.54281 |
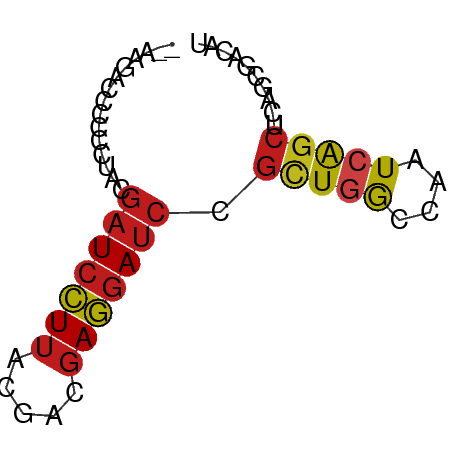
| Mean single sequence MFE | -15.06 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
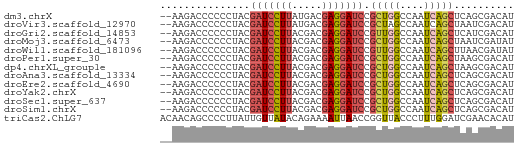
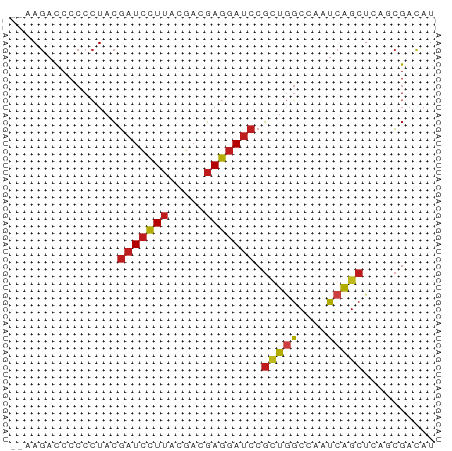
>dm3.chrX 16867008 57 - 22422827 --AAGACCCCCCUACGAUCCUUAUGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.47, R) >droVir3.scaffold_12970 9532111 57 - 11907090 --AAGACCCCCCUACGAUCCUUAUGACGAGGAUCCGCUAGCCAAUCAGCUAAUCGACAU --.............(((((((.....)))))))((.((((......))))..)).... ( -11.60, z-score = -2.24, R) >droGri2.scaffold_14853 9683441 57 + 10151454 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGUUGGCCAAUCAGCUCAUCGACAU --.............(((((((.....)))))))((.((((......)).)).)).... ( -11.80, z-score = -1.02, R) >droMoj3.scaffold_6473 4883533 57 + 16943266 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUAAUCGAUAU --.............(((((((.....))))))).(((((....))))).......... ( -13.50, z-score = -1.99, R) >droWil1.scaffold_181096 6093857 57 - 12416693 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGUUGGCCAAUCAGCUUAACGAUAU --.............(((((((.....)))))))(((((((......)).))))).... ( -13.60, z-score = -2.35, R) >droPer1.super_30 757478 57 + 958394 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUAAGCGACAU --.............(((((((.....)))))))(((((((......))).)))).... ( -16.00, z-score = -3.05, R) >dp4.chrXL_group1e 995709 57 - 12523060 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUAAGCGACAU --.............(((((((.....)))))))(((((((......))).)))).... ( -16.00, z-score = -3.05, R) >droAna3.scaffold_13334 650195 57 + 1562580 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.65, R) >droEre2.scaffold_4690 7220381 57 - 18748788 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.65, R) >droYak2.chrX 15475760 57 - 21770863 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.65, R) >droSec1.super_637 4369 57 - 9632 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.65, R) >droSim1.chrX 13048246 57 - 17042790 --AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU --.............(((((((.....)))))))(((((((......)).))))).... ( -18.30, z-score = -3.65, R) >triCas2.ChLG7 11679078 59 - 17478683 ACAACAGCCCCUUAUUGUUAUACAGAAAAUUAACCGGUUACCCUUUGGAUCGAACACAU ..(((((.......)))))...............(((((........)))))....... ( -3.50, z-score = 1.18, R) >consensus __AAGACCCCCCUACGAUCCUUACGACGAGGAUCCGCUGGCCAAUCAGCUCAGCGACAU ...............(((((((.....))))))).(((((....))))).......... (-10.76 = -10.98 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:51:00 2011