| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,858,022 – 16,858,120 |
| Length | 98 |
| Max. P | 0.971864 |

| Location | 16,858,022 – 16,858,120 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.59 |
| Shannon entropy | 0.66276 |
| G+C content | 0.39417 |
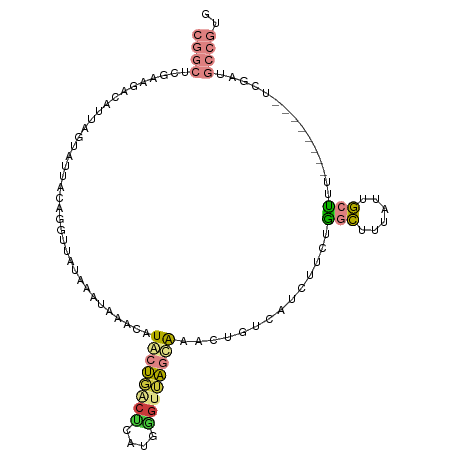
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.35 |
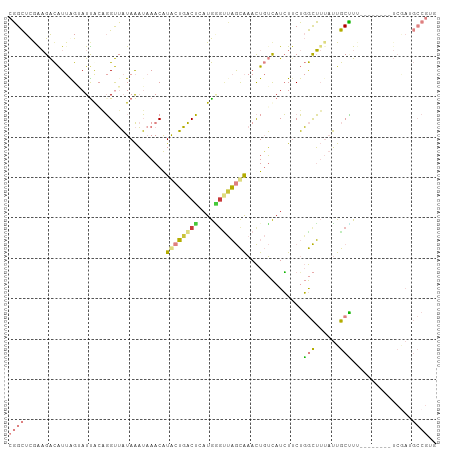
| Combinations/Pair | 1.67 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
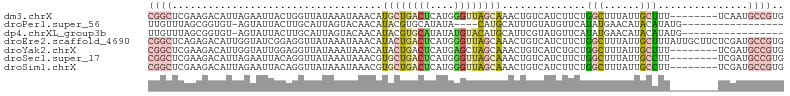
>dm3.chrX 16858022 98 + 22422827 CGGCUCGAAGACAUUAGAAUUACUGGUUAUAAAUAAACAUGCUGACUCAUGGGUUAGCAAACUGUCAUCUUCUGGCUUUAUUGCUUU--------UCAAUGCCGUG ((((..(((((...(((........(((.......))).((((((((....))))))))..)))...))))).(((......)))..--------.....)))).. ( -25.20, z-score = -2.39, R) >droPer1.super_56 203046 84 - 431378 UUGUUUAGCGGUGU-AGUAUUACUUGCAUUAGUACAACAUACGUGCAUAUA----CAUGCAUUUGUAUGUUCAUAUGAACAUACAUAUG----------------- .......(((((((-.((((((.......)))))).)))).)))((((...----.))))...(((((((((....)))))))))....----------------- ( -22.60, z-score = -1.39, R) >dp4.chrXL_group3b 183992 88 - 386183 UUGUUUAGCGGUGU-AGUAUUACUUGCAUUAGUACAACAUACGUGCAUAUAUGUACAUGCAUUCGUAUGUUCAUAUGAACAUACAUAUG----------------- .......(((((((-.((((((.......)))))).))))..(((((....))))).)))....((((((((....)))))))).....----------------- ( -23.90, z-score = -1.23, R) >droEre2.scaffold_4690 7212237 106 + 18748788 CGGCUCAGAGACAUUGGUAUCGGAGGUUAUAAAUAAACAUACUGACUCAUGGGUUAGCAAACUGUCAUCUUCUGGCUUUAUUGCUUUAUUGCUUCUCGAUGCCGUG ...............((((((((((((.(((((.........(((((....)))))((((...((((.....))))....))))))))).))))).)))))))... ( -28.60, z-score = -1.96, R) >droYak2.chrX 15467803 98 + 21770863 CGGCUCGAAGACAUUGGUAUUGGAGGUUAUAAAUAAACAUACUGACUCAUGAGCUAGCAAACUGUCAUCUGCUGGCUUUAUUGCUUU--------UCGAUGCCGUG ((((((((((((((..((((.(...((.....))...)))))..).....(((((((((..........)))))))))...)).)))--------)))).)))).. ( -27.80, z-score = -2.26, R) >droSec1.super_17 691456 98 + 1527944 CGGCUCGAAGACAUUAGAAUUACAGGUUAUAAAUAAACGUGCUGACUCAUGGGUUAGCAAACUGUCAUCUUCUGGCUUUAUUGCCUU--------UCGAUGCCGUG ((((((((((.....((((..(((((((.......))).((((((((....))))))))..))))....))))(((......)))))--------)))).)))).. ( -32.20, z-score = -3.84, R) >droSim1.chrX 13038714 98 + 17042790 CGGCUCGAAGACAUUAGAAUUACAGGUUAUAAAUAAACGUGCUGACUCAUGGGUUAGCAAACUGUCAUCUUCUGGCUUUAUUGCCUU--------UCGAUGCCGUG ((((((((((.....((((..(((((((.......))).((((((((....))))))))..))))....))))(((......)))))--------)))).)))).. ( -32.20, z-score = -3.84, R) >consensus CGGCUCGAAGACAUUAGUAUUACAGGUUAUAAAUAAACAUACUGACUCAUGGGUUAGCAAACUGUCAUCUUCUGGCUUUAUUGCUUU________UCGAUGCCGUG ((((...................................((((((((....))))))))..............(((......)))...............)))).. ( -8.82 = -9.17 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:59 2011