
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,836,551 – 16,836,622 |
| Length | 71 |
| Max. P | 0.990939 |

| Location | 16,836,551 – 16,836,622 |
|---|---|
| Length | 71 |
| Sequences | 11 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 87.28 |
| Shannon entropy | 0.27168 |
| G+C content | 0.52196 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -24.18 |
| Energy contribution | -23.54 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
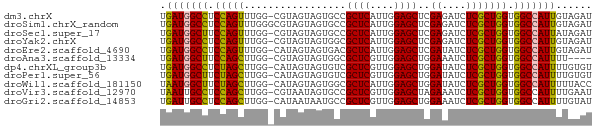
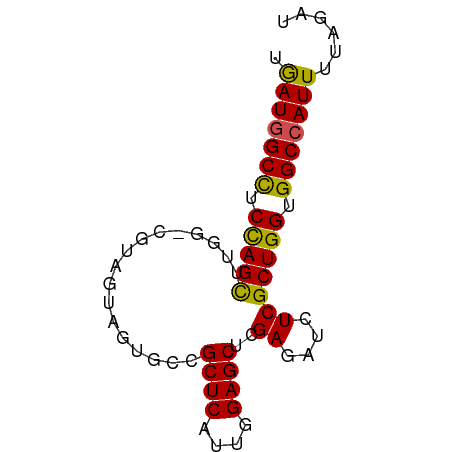
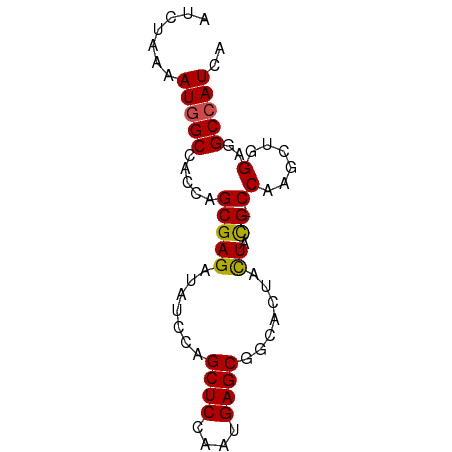
>dm3.chrX 16836551 71 + 22422827 UGAUGGCCUCCAGUUUGG-CGUAGUAGUGCCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUGUAGAU .(((((((.(((((..((-((......))))((((....))))..........))))).)))))))...... ( -30.50, z-score = -2.88, R) >droSim1.chrX_random 4373015 72 + 5698898 UGAUGGCCUCCAGUUUGGGCGUAGUAGUGCCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUGUAGAU .(((((((.(((((..(((((......))))((((....)))).......)..))))).)))))))...... ( -30.80, z-score = -2.62, R) >droSec1.super_17 670243 71 + 1527944 UGAUGGCUUCCAGUUUGG-CGUAGUAGUGCCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUAUAGAU ((((((((.(((((..((-((......))))((((....))))..........))))).))))))))..... ( -28.60, z-score = -2.59, R) >droYak2.chrX 15446848 71 + 21770863 UGAUGGCCUCCAGUUUGG-CGUAGUAGUGGCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUGUAGAU .(((((((.(((.....(-((..((..(((.((((....)))))))..))..)))))).)))))))...... ( -27.00, z-score = -1.56, R) >droEre2.scaffold_4690 7190400 71 + 18748788 UGAUGGCCUCCAGUUUGG-CAUAGUAGUGACGCUCAUUGGAGCUCGAUAUCUCGCUGGUGGCCAUUGUAGAU .(((((((.(((((..((-.(((....(((.((((....))))))).))))).))))).)))))))...... ( -27.90, z-score = -2.53, R) >droAna3.scaffold_13334 976533 67 - 1562580 UGAUGGCUUCCAGCUUGG-CGUAGUAGUGGCGCUCGUUGGAGCUGGAAAUCUCGCUGGUGGCCAUUUU---- .(((((((.(((((..((-(((.......))))).....(((.(....).)))))))).)))))))..---- ( -27.10, z-score = -2.16, R) >dp4.chrXL_group3b 152235 71 - 386183 UGAUGGCCUCUAGCUUGG-CAUAGUAGUGUCGCUCGUUGGAGCUGGAUAUCUCGCUGGUGGCCAUUUUGUGU .(((((((.(((((..((-(((....)))))((((....))))..........))))).)))))))...... ( -29.60, z-score = -3.13, R) >droPer1.super_56 171143 71 - 431378 UGAUGGCUUCUAGCUUGG-CAUAGUAGUGUCGCUCGUUGGAGCUGGAUAUCUCGCUGGUGGCCAUUUUGUGU .(((((((.(((((..((-(((....)))))((((....))))..........))))).)))))))...... ( -27.00, z-score = -2.59, R) >droWil1.scaffold_181150 328952 71 - 4952429 UAAUGGCUUCUAGCUUGG-CAUAGUAGUGGCGCUCAUUGGAGCUGGAUAUCUCGCUGGUGGCCAUUUUUACC .(((((((.(((((..(.-(((....))).)((((....))))..........))))).)))))))...... ( -24.50, z-score = -1.68, R) >droVir3.scaffold_12970 5413775 71 - 11907090 UAAUUGCCUCCAGCUUGG-CGUAAUAGUGCCGCUCGUUGGAGCUAGAAAUCUCGCUGGUGGCCAUUUUGAAU .....(((.(((((..((-((......))))((((....))))..........))))).))).......... ( -25.80, z-score = -2.57, R) >droGri2.scaffold_14853 5998449 71 + 10151454 UGAUUGCCUCCAGCUUGG-CAUAAUAAUGCCGCUCGUUGGAGCUGGAAAUCUCGCUGGUGGCCAUUUUGUAU .(((.(((.(((((..((-(((....)))))((((....))))..........))))).))).)))...... ( -27.80, z-score = -3.08, R) >consensus UGAUGGCCUCCAGCUUGG_CGUAGUAGUGCCGCUCAUUGGAGCUCGAGAUCUCGCUGGUGGCCAUUUUAGAU .(((((((.(((((.................((((....))))..((....))))))).)))))))...... (-24.18 = -23.54 + -0.64)
| Location | 16,836,551 – 16,836,622 |
|---|---|
| Length | 71 |
| Sequences | 11 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 87.28 |
| Shannon entropy | 0.27168 |
| G+C content | 0.52196 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -9.08 |
| Energy contribution | -9.26 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
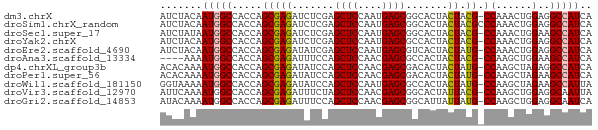
>dm3.chrX 16836551 71 - 22422827 AUCUACAAUGGCCACCAGCGAGAUCUCGAGCUCCAAUGAGCGGCACUACUACG-CCAAACUGGAGGCCAUCA .......((((((.(((((((....))).((((....))))(((........)-))...)))).)))))).. ( -27.40, z-score = -4.09, R) >droSim1.chrX_random 4373015 72 - 5698898 AUCUACAAUGGCCACCAGCGAGAUCUCGAGCUCCAAUGAGCGGCACUACUACGCCCAAACUGGAGGCCAUCA .......((((((.(((((((....))).((((....))))(((........)))....)))).)))))).. ( -26.50, z-score = -3.67, R) >droSec1.super_17 670243 71 - 1527944 AUCUAUAAUGGCCACCAGCGAGAUCUCGAGCUCCAAUGAGCGGCACUACUACG-CCAAACUGGAAGCCAUCA .......(((((..(((((((....))).((((....))))(((........)-))...))))..))))).. ( -23.20, z-score = -3.10, R) >droYak2.chrX 15446848 71 - 21770863 AUCUACAAUGGCCACCAGCGAGAUCUCGAGCUCCAAUGAGCGCCACUACUACG-CCAAACUGGAGGCCAUCA .......((((((.(((((((....))).((((....))))............-.....)))).)))))).. ( -23.20, z-score = -3.07, R) >droEre2.scaffold_4690 7190400 71 - 18748788 AUCUACAAUGGCCACCAGCGAGAUAUCGAGCUCCAAUGAGCGUCACUACUAUG-CCAAACUGGAGGCCAUCA .......((((((.(((((((....))).((((....))))............-.....)))).)))))).. ( -23.10, z-score = -2.94, R) >droAna3.scaffold_13334 976533 67 + 1562580 ----AAAAUGGCCACCAGCGAGAUUUCCAGCUCCAACGAGCGCCACUACUACG-CCAAGCUGGAAGCCAUCA ----...(((((..(((((..........((((....))))((.........)-)...)))))..))))).. ( -20.80, z-score = -3.10, R) >dp4.chrXL_group3b 152235 71 + 386183 ACACAAAAUGGCCACCAGCGAGAUAUCCAGCUCCAACGAGCGACACUACUAUG-CCAAGCUAGAGGCCAUCA .......((((((.(.(((((....))..((((....))))............-....))).).)))))).. ( -18.50, z-score = -2.78, R) >droPer1.super_56 171143 71 + 431378 ACACAAAAUGGCCACCAGCGAGAUAUCCAGCUCCAACGAGCGACACUACUAUG-CCAAGCUAGAAGCCAUCA .......(((((..(.(((((....))..((((....))))............-....))).)..))))).. ( -14.30, z-score = -1.92, R) >droWil1.scaffold_181150 328952 71 + 4952429 GGUAAAAAUGGCCACCAGCGAGAUAUCCAGCUCCAAUGAGCGCCACUACUAUG-CCAAGCUAGAAGCCAUUA ......((((((..(.(((((....))..((((....))))............-....))).)..)))))). ( -15.70, z-score = -0.84, R) >droVir3.scaffold_12970 5413775 71 + 11907090 AUUCAAAAUGGCCACCAGCGAGAUUUCUAGCUCCAACGAGCGGCACUAUUACG-CCAAGCUGGAGGCAAUUA ..........(((.(((((..........((((....))))(((........)-))..))))).)))..... ( -25.90, z-score = -3.89, R) >droGri2.scaffold_14853 5998449 71 - 10151454 AUACAAAAUGGCCACCAGCGAGAUUUCCAGCUCCAACGAGCGGCAUUAUUAUG-CCAAGCUGGAGGCAAUCA ..........(((.(((((..........((((....))))(((((....)))-))..))))).)))..... ( -28.40, z-score = -4.79, R) >consensus AUCUAAAAUGGCCACCAGCGAGAUAUCCAGCUCCAAUGAGCGGCACUACUACG_CCAAGCUGGAGGCCAUCA .......(((((.......((....))..((((....))))........................))))).. ( -9.08 = -9.26 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:55 2011