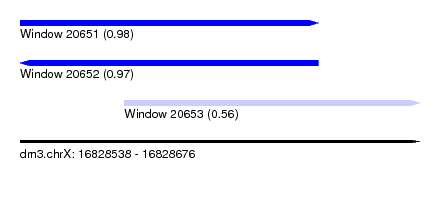
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,828,538 – 16,828,676 |
| Length | 138 |
| Max. P | 0.977246 |

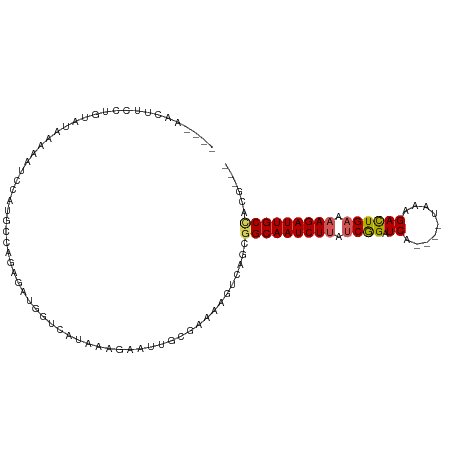
| Location | 16,828,538 – 16,828,641 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Shannon entropy | 0.37087 |
| G+C content | 0.39472 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
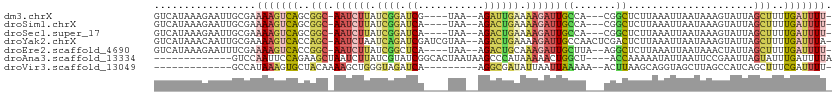
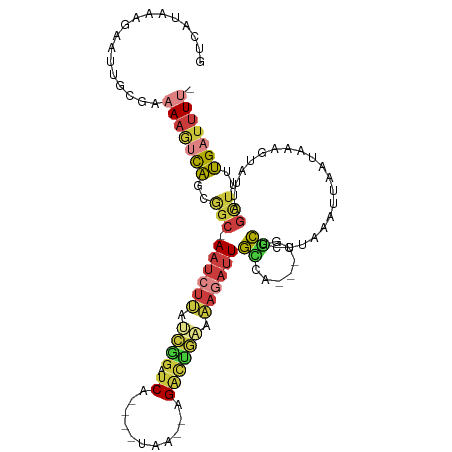
>dm3.chrX 16828538 103 + 22422827 CUCGCGCUUCCUUAAUAAAAAUCCACUCCAAAGAUGGUCAUAAAGAAUUGCGAAAAGUCAGCGGCAAUCUUAUCGGAUCG----UAAAGAUUGAAAAGAUUGCCACG--- .(((((.(((.(((........(((((....)).)))...))).))).))))).......(.(((((((((.(((.(((.----....)))))).))))))))).).--- ( -26.30, z-score = -2.68, R) >droSim1.chrX 13019150 99 + 17042790 ----AACUUCCUGUAUAAAAUUCCAUGCCAGAGAUGGUCAUAAAGAAUUGCGAAAAGUCAGCGGCAAUCUUAUCGGAUCA----UAAAGACUGAAAAGAUUGCCACG--- ----.((((..((((....((((.((((((....))).)))...))))))))..))))..(.(((((((((.((((.((.----....)))))).))))))))).).--- ( -27.30, z-score = -3.33, R) >droSec1.super_17 662613 99 + 1527944 ----AACUUCCUGUAUAAAAUUCCAUGCCAGAGAUGGUCAUAAAGAAUUGCGAAAAGUCAGCGGCAAUCUUAUCGGAUCA----UAAAGACUGAAAAGAUUGCCACG--- ----.((((..((((....((((.((((((....))).)))...))))))))..))))..(.(((((((((.((((.((.----....)))))).))))))))).).--- ( -27.30, z-score = -3.33, R) >droYak2.chrX 15439183 98 + 21770863 --------CACUCUCGAAAUGGUCAUAACUGAG----UCAUAAACAAUUGCGAAAAGUCACCAGCAAUCUAAUCAGAUCGAUCGUAAAGACUGAAAAGAUUGCCAACUCG --------......(((..((((....(((...----((.(((....))).))..))).))))(((((((..((((.((.........))))))..)))))))....))) ( -18.30, z-score = -1.15, R) >droEre2.scaffold_4690 7183190 100 + 18748788 ----CACUUCCUUUAUAAAAAUCCACUCCAGAGAUGGUCAUAAAGAAUUUCGAAAAGUCACCGGCAAUCUUAUCGGCUCA----UAAAGACUGCAAAGAUUGCUUAAG-- ----......((((((......(((((....)).)))..)))))).................(((((((((..(((.((.----....)))))..)))))))))....-- ( -17.50, z-score = -1.00, R) >consensus ____AACUUCCUGUAUAAAAAUCCAUGCCAGAGAUGGUCAUAAAGAAUUGCGAAAAGUCAGCGGCAAUCUUAUCGGAUCA____UAAAGACUGAAAAGAUUGCCACG___ ................................(((..((.(((....))).))...)))...(((((((((.((((.((.........)))))).)))))))))...... (-15.80 = -16.56 + 0.76)

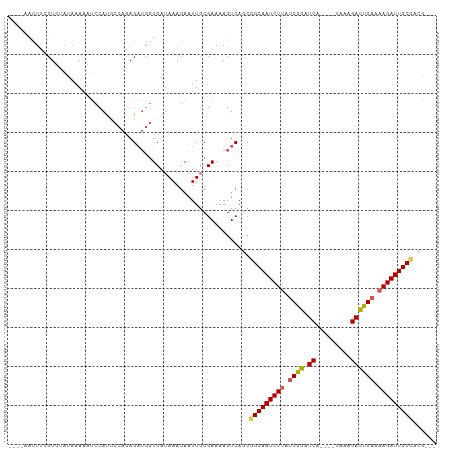
| Location | 16,828,538 – 16,828,641 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Shannon entropy | 0.37087 |
| G+C content | 0.39472 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -16.42 |
| Energy contribution | -18.50 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16828538 103 - 22422827 ---CGUGGCAAUCUUUUCAAUCUUUA----CGAUCCGAUAAGAUUGCCGCUGACUUUUCGCAAUUCUUUAUGACCAUCUUUGGAGUGGAUUUUUAUUAAGGAAGCGCGAG ---.(((((((((((.((.(((....----.)))..)).))))))))))).......((((..((((((((((..((((.......))))..))).)))))))..)))). ( -32.70, z-score = -3.65, R) >droSim1.chrX 13019150 99 - 17042790 ---CGUGGCAAUCUUUUCAGUCUUUA----UGAUCCGAUAAGAUUGCCGCUGACUUUUCGCAAUUCUUUAUGACCAUCUCUGGCAUGGAAUUUUAUACAGGAAGUU---- ---.(((((((((((.((.(((....----.)))..)).))))))))))).((((((..(.((((((..(((.(((....)))))))))))).....)..))))))---- ( -29.90, z-score = -3.48, R) >droSec1.super_17 662613 99 - 1527944 ---CGUGGCAAUCUUUUCAGUCUUUA----UGAUCCGAUAAGAUUGCCGCUGACUUUUCGCAAUUCUUUAUGACCAUCUCUGGCAUGGAAUUUUAUACAGGAAGUU---- ---.(((((((((((.((.(((....----.)))..)).))))))))))).((((((..(.((((((..(((.(((....)))))))))))).....)..))))))---- ( -29.90, z-score = -3.48, R) >droYak2.chrX 15439183 98 - 21770863 CGAGUUGGCAAUCUUUUCAGUCUUUACGAUCGAUCUGAUUAGAUUGCUGGUGACUUUUCGCAAUUGUUUAUGA----CUCAGUUAUGACCAUUUCGAGAGUG-------- ....(..(((((((..((((((.........)).))))..)))))))..)..(((((.((.((.((.((((((----(...))))))).)).))))))))).-------- ( -26.70, z-score = -1.77, R) >droEre2.scaffold_4690 7183190 100 - 18748788 --CUUAAGCAAUCUUUGCAGUCUUUA----UGAGCCGAUAAGAUUGCCGGUGACUUUUCGAAAUUCUUUAUGACCAUCUCUGGAGUGGAUUUUUAUAAAGGAAGUG---- --.....((((((((.((..((....----.))))....)))))))).(....).........(((((((((((((.((....)))))....))))))))))....---- ( -20.20, z-score = 0.04, R) >consensus ___CGUGGCAAUCUUUUCAGUCUUUA____UGAUCCGAUAAGAUUGCCGCUGACUUUUCGCAAUUCUUUAUGACCAUCUCUGGAAUGGAAUUUUAUAAAGGAAGUG____ ....(((((((((((.((.(((.........)))..)).))))))))))).............((((((((((((((.......))))....))))))))))........ (-16.42 = -18.50 + 2.08)

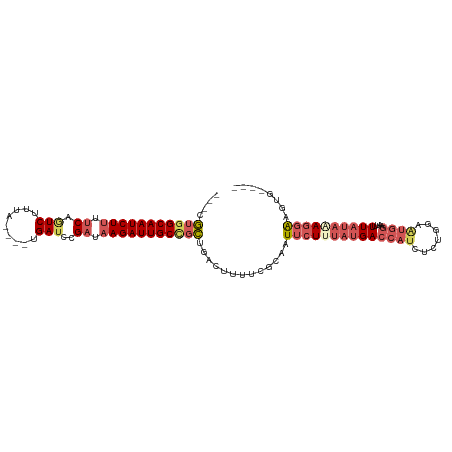
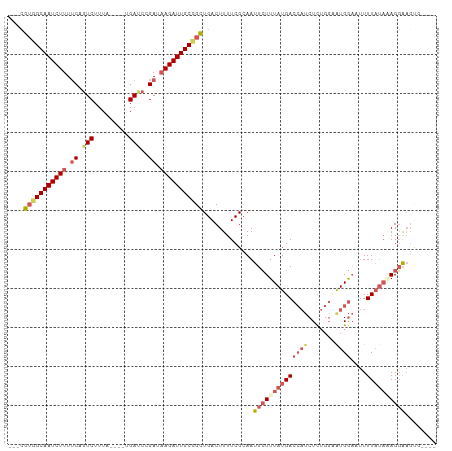
| Location | 16,828,574 – 16,828,676 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.99 |
| Shannon entropy | 0.64399 |
| G+C content | 0.33420 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -6.91 |
| Energy contribution | -7.94 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
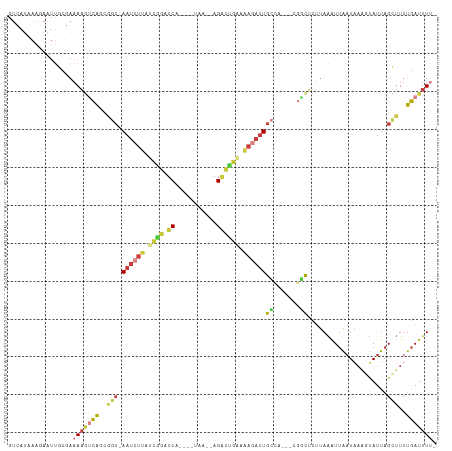
>dm3.chrX 16828574 102 + 22422827 GUCAUAAAGAAUUGCGAAAAGUCAGCGGC-AAUCUUAUCGGAUCG----UAA--AGAUUGAAAAGAUUGCCA---CGGCUCUUAAAUUAAUAAAGUAUUAGCUUUUGAUUUU- ((((.((((...(((.((.((((.(.(((-((((((.(((.(((.----...--.)))))).))))))))).---))))).))...........)))....))))))))...- ( -24.40, z-score = -2.10, R) >droSim1.chrX 13019182 102 + 17042790 GUCAUAAAGAAUUGCGAAAAGUCAGCGGC-AAUCUUAUCGGAUCA----UAA--AGACUGAAAAGAUUGCCA---CGGCUCUUAAAUUAAUAAAGUAUUAGCUUUUGAUUUU- ((((.((((...(((.((.((((.(.(((-((((((.((((.((.----...--.)))))).))))))))).---))))).))...........)))....))))))))...- ( -25.60, z-score = -2.75, R) >droSec1.super_17 662645 102 + 1527944 GUCAUAAAGAAUUGCGAAAAGUCAGCGGC-AAUCUUAUCGGAUCA----UAA--AGACUGAAAAGAUUGCCA---CGGCUCUUAAAUUAAUAAAGUAUUAGCUUUUGAUUUU- ((((.((((...(((.((.((((.(.(((-((((((.((((.((.----...--.)))))).))))))))).---))))).))...........)))....))))))))...- ( -25.60, z-score = -2.75, R) >droYak2.chrX 15439207 109 + 21770863 GUCAUAAACAAUUGCGAAAAGUCACCAGC-AAUCUAAUCAGAUCGAUCGUAA--AGACUGAAAAGAUUGCCAACUCGACUCUUAAAUUAAUAAAGUAUUAGCUUUUGAUUUA- ........(((..((....((((....((-(((((..((((.((........--.))))))..)))))))......)))).......(((((...)))))))..))).....- ( -19.60, z-score = -1.65, R) >droEre2.scaffold_4690 7183222 103 + 18748788 GUCAUAAAGAAUUUCGAAAAGUCACCGGC-AAUCUUAUCGGCUCA----UAA--AGACUGCAAAGAUUGCUUA--AGGCUCUUAAAUUAAUAAACUAUUAGCUUUUGAUUUU- .................(((((((..(((-((((((..(((.((.----...--.)))))..))))))))).(--(((((..((...........))..)))))))))))))- ( -19.00, z-score = -1.06, R) >droAna3.scaffold_13334 968404 96 - 1562580 -------------GUCCAAUUCCAGAAGCUAAUCUUAUCGUAUCGGCACUAAUAAGCCCAUAAAAACUGGCU----ACCAAAAAUAUUAAUUCCGAAUUAGUAUUUGAUUUUA -------------.........((((.((((((.........((((...(((((((((..........))))----........)))))...)))))))))).))))...... ( -13.20, z-score = -0.95, R) >droVir3.scaffold_13049 12711650 88 + 25233164 -------------GCCAUAAAGUGCUACAAAAGCUGGGUAGAUCA---------AGGCGAUAUUAAUUAAAAA--ACUUAAGCAGGUAGCUUAGCCAUCAGCUUUCGAUUUU- -------------..............(.((((((((((..(((.---------....)))............--...(((((.....))))))))..))))))).).....- ( -15.00, z-score = 0.74, R) >consensus GUCAUAAAGAAUUGCGAAAAGUCAGCGGC_AAUCUUAUCGGAUCA____UAA__AGACUGAAAAGAUUGCCA___CGGCUCUUAAAUUAAUAAAGUAUUAGCUUUUGAUUUU_ .................(((((((..(((.((((((.((((.((...........)))))).))))))(((.....))).....................)))..))))))). ( -6.91 = -7.94 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:53 2011