
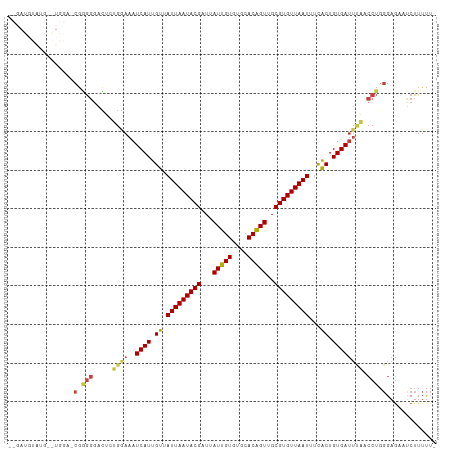
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,815,272 – 16,815,405 |
| Length | 133 |
| Max. P | 0.999236 |

| Location | 16,815,272 – 16,815,383 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Shannon entropy | 0.42592 |
| G+C content | 0.37948 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16815272 111 + 22422827 UUGAUGUAUGUGUGGCCCGGGGGACUCUGGAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUUCACUGUGAUUUAACCUGGGAGAAUCUUUUUU ...............((((((.........(((((((((..(((((((((...(((((....)))))..)))))))))..))..)))))))..))))))............ ( -29.30, z-score = -1.59, R) >droYak2.chrX 15425733 109 + 21770863 ACGAUGUAUGAAUGGCCCGGGG-ACUGUGGAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUCCACUGUGAUUUAACCUGGGAGAAUCUUUUU- .........((.(..((((((.-.......(((((((((..(((((((((...(((((....)))))..)))))))))..))..)))))))..))))))..).)).....- ( -30.20, z-score = -1.90, R) >droSec1.super_17 649591 111 + 1527944 UCGAUGUAUGUGUGGAACGGGGGACUCUGGAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUUCACUGUGAUUUAACCUGGGAGAAUCUUUUUU ..............(((..(((..(((((((((((((((..(((((((((...(((((....)))))..)))))))))..))..)))))))..))..))))..)))..))) ( -26.10, z-score = -0.87, R) >droSim1.chrX 13005402 111 + 17042790 UUGAUGUAUGUGUGGAACGGGGGACUCUGGAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUUCACUGUGAUUUAACCUGGGAGAAUCUUUUUU ..............(((..(((..(((((((((((((((..(((((((((...(((((....)))))..)))))))))..))..)))))))..))..))))..)))..))) ( -26.10, z-score = -1.12, R) >dp4.chrXL_group3b 131855 93 - 386183 -----------------CGAGGGAUGGGGAAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACGGUUGCGUGUUAAUUUUGCUGUGAUUCCACCUGGGAGUACCUUUUU- -----------------.((((..((((...((((((.((.(((((((((...(((((....)))))..)))))))))...)).))))))...))))......))))...- ( -25.00, z-score = -1.55, R) >droPer1.super_56 150814 93 - 431378 -----------------CGAGGGAUGGGGAAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACGGUUGCGUGUUAAUUUUGCUGUGAUUCCACCUGGGAGUACCUUUUU- -----------------.((((..((((...((((((.((.(((((((((...(((((....)))))..)))))))))...)).))))))...))))......))))...- ( -25.00, z-score = -1.55, R) >droAna3.scaffold_13334 954366 84 - 1562580 ---------------AUUAUAUGAGACACAAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUUCGCUGUGA----------GAAUUUCUUUC-- ---------------.......((((.......((((.((.(((((((((...(((((....)))))..)))))))))...)).))))----------.....))))..-- ( -17.40, z-score = -1.01, R) >consensus __GAUGUAUG__UGGA_CGGGGGACUCUGGAAAUCAUUGUUAUUAAUACGAUUAUUGUGUGCACAGUUGCGUGUUAAUUUCACUGUGAUUUAACCUGGGAGAAUCUUUUU_ .................(.(((.....((((..((((.((.(((((((((...(((((....)))))..)))))))))...)).)))))))).))).)............. (-18.52 = -18.40 + -0.12)


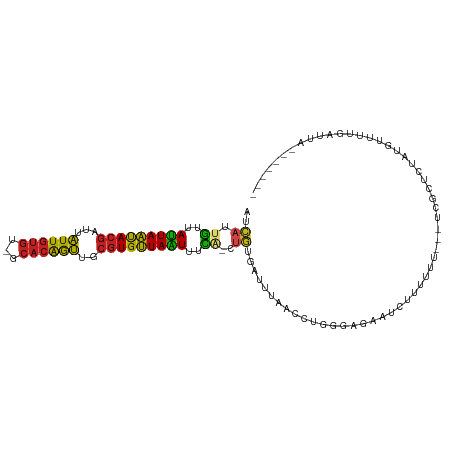
| Location | 16,815,304 – 16,815,405 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.65258 |
| G+C content | 0.33367 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -13.13 |
| Energy contribution | -12.41 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16815304 101 + 22422827 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUCA-CUGUGAUUUAACCUGGGAGAAUCUUUUUUGUCUCGCUCUAUGUUUUGAUUA------- (((((((..(((((((((...(((((..-..)))))..)))))))))..))-..))))).......((((.((......)).)))).................------- ( -21.20, z-score = -1.66, R) >droEre2.scaffold_4690 7170719 99 + 18748788 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUCA-CUGUGAUUUAACCUGGGAGAAUCUUUUUU--CUCGCUCUAUGUUUUGACUA------- (((((((..(((((((((...(((((..-..)))))..)))))))))..))-..))))).......(.(((((......))--))).)...............------- ( -22.90, z-score = -2.51, R) >droYak2.chrX 15425764 99 + 21770863 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUCCA-CUGUGAUUUAACCUGGGAGAAUCUUUUUU--CUCCCUCUAUGUUUUGAUUA------- (((((((..(((((((((...(((((..-..)))))..)))))))))..))-..))))).......(((((((......))--)))))...............------- ( -28.10, z-score = -4.97, R) >droSec1.super_17 649623 101 + 1527944 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUCA-CUGUGAUUUAACCUGGGAGAAUCUUUUUUUUCUCGCUCUAUGUUUUGAUUA------- (((((((..(((((((((...(((((..-..)))))..)))))))))..))-..))))).......(((((((......))))))).................------- ( -22.70, z-score = -2.60, R) >droSim1.chrX 13005434 101 + 17042790 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUCA-CUGUGAUUUAACCUGGGAGAAUCUUUUUUUUCUCGCUCUAUGUUUUGAUUA------- (((((((..(((((((((...(((((..-..)))))..)))))))))..))-..))))).......(((((((......))))))).................------- ( -22.70, z-score = -2.60, R) >dp4.chrXL_group3b 131870 95 - 386183 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACGGUUGCGUGUUAAUUUUG-CUGUGAUUCCACCUGGGAGUACCUUUUU------GUUUGGCGUUUUUCCCA------- (((((.((.(((((((((...(((((..-..)))))..)))))))))...)-).))))).......(((((.((((....------....)).))..))))).------- ( -24.00, z-score = -2.02, R) >droPer1.super_56 150829 95 - 431378 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACGGUUGCGUGUUAAUUUUG-CUGUGAUUCCACCUGGGAGUACCUUUUU------GUUUGGCGUUUUUCCCA------- (((((.((.(((((((((...(((((..-..)))))..)))))))))...)-).))))).......(((((.((((....------....)).))..))))).------- ( -24.00, z-score = -2.02, R) >droAna3.scaffold_13334 954383 88 - 1562580 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUCG-CUGUGA----------GAAUUUCUUUCCU---CCACACUAUAUAUAUACUA------- .((((.((.(((((((((...(((((..-..)))))..)))))))))...)-).))))----------.............---...................------- ( -16.40, z-score = -1.49, R) >droWil1.scaffold_181150 4649381 100 + 4952429 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAGUUUU--CUGUGAUUUUUGCCCACAUAAUCUAUUCUUUGUUUUGUUACUUUUUUUUUC------- (((((.(..(((((((((...(((((..-..)))))..)))))))))...--).)))))............................................------- ( -15.80, z-score = -2.02, R) >droVir3.scaffold_12970 6516824 105 + 11907090 AUCAUUGUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUUU-CUGUAAUUUUCGCCCAGAGAUUGUUGUUU---UAGUUUUCCCAUUUUUUUUAUUUUGU .........(((((((((...(((((..-..)))))..))))))))).(((-(((...........)))))).........---.......................... ( -14.90, z-score = -1.53, R) >droMoj3.scaffold_6473 15469450 91 - 16943266 --GUUUAUUAUUAAUACGAUUAUUGUGU-GCACAGUUGCGUGUUAAUUUUU-CUGUAAUUUGUGCCCAAAGUUUGUUGUUU---UAGUUUUUUUUUUA------------ --.......(((((.(((((.(((.((.-(((((((((((.(........)-.))))).)))))).)).)))..))))).)---))))..........------------ ( -15.60, z-score = -2.25, R) >droGri2.scaffold_14853 5976479 109 + 10151454 GUUCAUGCGAAUAGCACGGCUUUUCCAUAGCCAAAAUAACUGCUAUUUUCAUUUAUUGUUCGCGUUCUUCGAGUGCUUCGCGUGACUUUCGGGCCAUAAACUACUGUGC- ..((((((((..(((((((((.......))))((((((.....)))))).......................))))))))))))).........((((......)))).- ( -24.90, z-score = -0.56, R) >consensus AUCAUUGUUAUUAAUACGAUUAUUGUGU_GCACAGUUGCGUGUUAAUUUCA_CUGUGAUUUAACCUGGGAGAAUCUUUUUU___UCGCUCUAUGUUUUGAUUA_______ ......(..(((((((((...((((((...))))))..)))))))))..)............................................................ (-13.13 = -12.41 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:50 2011