| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,770,545 – 16,770,599 |
| Length | 54 |
| Max. P | 0.927383 |

| Location | 16,770,545 – 16,770,599 |
|---|---|
| Length | 54 |
| Sequences | 11 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.63088 |
| G+C content | 0.54685 |
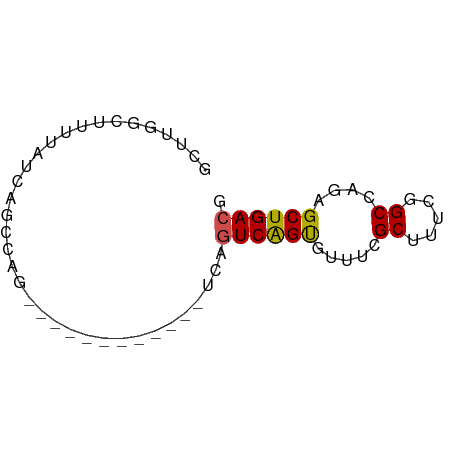
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -9.28 |
| Energy contribution | -9.15 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927383 |
| Prediction | RNA |
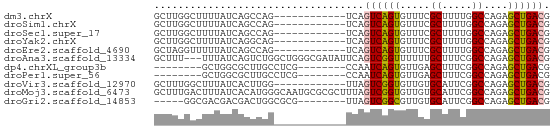
Download alignment: ClustalW | MAF
>dm3.chrX 16770545 54 - 22422827 GCUUGGCUUUUAUCAGCCAG------------UCAGUCAGUGUUUCGCUUUUGGCCAGAGCUGACG (.((((((......))))))------------.).((((((.(((.((.....)).))))))))). ( -17.20, z-score = -1.72, R) >droSim1.chrX 12956690 54 - 17042790 GCUUGGCUUUUAUCAGCCAG------------UCAGUCAGUGUUUCGCUUUUGGCCAGAGCUGACG (.((((((......))))))------------.).((((((.(((.((.....)).))))))))). ( -17.20, z-score = -1.72, R) >droSec1.super_17 598037 54 - 1527944 GCUUGGCUUUUAUCAGCCAG------------UCAGUCAGUGUUUCGCUUUUGGCCAGAGCUGACG (.((((((......))))))------------.).((((((.(((.((.....)).))))))))). ( -17.20, z-score = -1.72, R) >droYak2.chrX 15382112 54 - 21770863 GCUUGGCUUUUAUCAGGCAG------------UCAGUCAGUGUUUCGCUUUUGGCCAGAGCUGACG ((((((......))))))..------------...((((((.(((.((.....)).))))))))). ( -16.50, z-score = -1.21, R) >droEre2.scaffold_4690 7127481 54 - 18748788 GCUAGGUUUUUAUCAGCCAG------------UCAGUCAGUGUUUCGCUUUUGGCCAGAGCUGACG (((.((((......))))))------------)..((((((.(((.((.....)).))))))))). ( -14.00, z-score = -1.18, R) >droAna3.scaffold_13334 903972 63 + 1562580 GCUUU---UUUAUCAGUCUGGCUGGGCGAUAUUCAGUCGGUUUUUUGCUUUCGGCCAGAGCUGACG .....---....(((((((((((((((((...((....))....)))))..)))))))).)))).. ( -21.70, z-score = -2.88, R) >dp4.chrXL_group3b 78698 50 + 386183 --------GCUGGCGCUUGCCUCG--------CCAAUCAGUGUUGAGCUUUCGGCCAGAGCUGACG --------..(((((.......))--------))).(((((.(((.((.....))))).))))).. ( -15.70, z-score = 0.04, R) >droPer1.super_56 95213 50 + 431378 --------GCUGGCGCUUGCCUCG--------CCAAUCAGUGUUGAGCUUUCGGCCAGAGCUGACG --------..(((((.......))--------))).(((((.(((.((.....))))).))))).. ( -15.70, z-score = 0.04, R) >droVir3.scaffold_12970 4896459 54 + 11907090 GCUUUGGCUUUAUCACUUGG------------UUAGUCGGUGUUGUGCAUUCGGCCAGAGCUGACG (((((((((..(((((....------------............))).))..)))))))))..... ( -16.59, z-score = -1.45, R) >droMoj3.scaffold_6473 4456190 66 + 16943266 GCUUUGACUUUAUCACAUGGGCAAUGCGCGCUUUAGUCGGUGUUGUGCAUUCGGCCAGAGCUGACG ((((((.((.......(((.(((((((.(((....).))))))))).)))..)).))))))..... ( -17.00, z-score = 0.53, R) >droGri2.scaffold_14853 5558334 53 - 10151454 -----GGCGACGACGACUGGCGCG--------UUAGUCGGCGUUGUGCAUUCGGCCAGAGCUGACG -----.((.((((((.(((((...--------...)))))))))))))..(((((....))))).. ( -21.70, z-score = -1.10, R) >consensus GCUUGGCUUUUAUCAGCCAG____________UCAGUCAGUGUUUCGCUUUCGGCCAGAGCUGACG ...................................((((((.....((.....))....)))))). ( -9.28 = -9.15 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:44 2011