| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,745,746 – 16,745,865 |
| Length | 119 |
| Max. P | 0.598107 |

| Location | 16,745,746 – 16,745,855 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Shannon entropy | 0.16000 |
| G+C content | 0.54240 |
| Mean single sequence MFE | -35.79 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
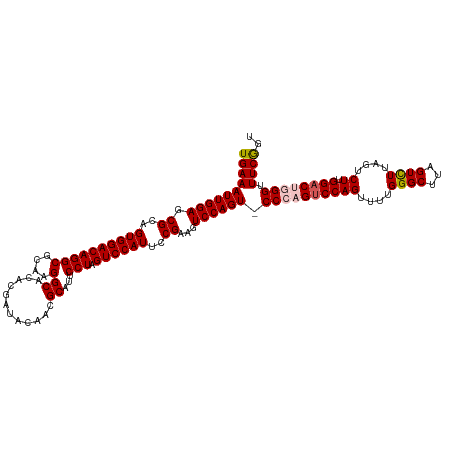
>dm3.chrX 16745746 109 + 22422827 -----------ACGAAACCCAGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGUACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGC -----------.....((((((((((......................)))))))))).(((..(....)..(((((.(((...(((..((((...))))..))).)))))))))))... ( -33.95, z-score = -1.00, R) >droSim1.chrX 12932480 110 + 17042790 ----------ACUGAAACCCGGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGCACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGC ----------.......((..(((((...(.(((.(((.(((((.........)))))..))).))).)...)))))..))...(((((((..((.((((...)))).))...))))))) ( -34.20, z-score = -0.77, R) >droSec1.super_17 573712 110 + 1527944 ----------ACUGAAACCCGGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGCACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACAGCGC ----------.......((..(((((...(.(((.(((.(((((.........)))))..))).))).)...)))))..))...(((((((..((.((((...)))).))...))))))) ( -35.00, z-score = -1.27, R) >droYak2.chrX 15357392 109 + 21770863 ----------ACCGAAACCCAGGCCGAGACUUAAACUAAGCCCAAAACUGGACUUGG-ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUAGCGCCUGUCCACUGCGC ----------.(((((..((((.(((((.((((...)))).(((....))).)))))-.))))..)))))..(((((.(((...((...((((...))))...)).))))))))...... ( -35.40, z-score = -2.22, R) >droEre2.scaffold_4690 7103414 119 + 18748788 CCAUACCGAUACCGAAACCCAGUCCGAGAUUAAGACUAAGCCCAAAACUGGACUUGG-ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACUGCGC ...........(((((..((((((((((.(((....)))..(((....))).)))))-)))))..)))))..(((((.(((...(((..((((...))))..))).))))))))...... ( -40.40, z-score = -3.55, R) >consensus __________ACCGAAACCCAGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGG_ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGC ...........(((((..(((((((.((.......)).((.(((....))).)).)).)))))..)))))..(((((.(((...(((..((((...))))..))).))))))))...... (-28.56 = -28.68 + 0.12)

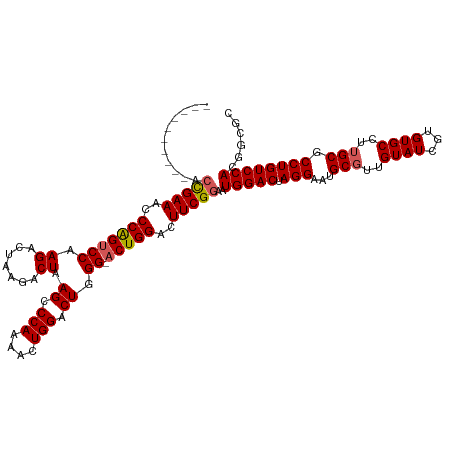
| Location | 16,745,746 – 16,745,865 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Shannon entropy | 0.09984 |
| G+C content | 0.52259 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -30.89 |
| Energy contribution | -31.21 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16745746 119 + 22422827 -ACGAAACCCAGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGUACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGCUCCAAUUUCA -.((..((((((((((......................))))))))))..))......((((((((..(((.(((((......))))).))).(((((.......)))))))).))))). ( -38.45, z-score = -1.65, R) >droSim1.chrX 12932480 120 + 17042790 ACUGAAACCCGGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGCACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGCUCCAAUUUCA ...((((((..(((((...(.(((.(((.(((((.........)))))..))).))).)...)))))..))...(((((((..((.((((...)))).))...))))))).....)))). ( -37.00, z-score = -0.87, R) >droSec1.super_17 573712 120 + 1527944 ACUGAAACCCGGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGGCACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACAGCGCUCCAAUUUCA ...((((((..(((((...(.(((.(((.(((((.........)))))..))).))).)...)))))..))...(((((((..((.((((...)))).))...))))))).....)))). ( -37.80, z-score = -1.40, R) >droYak2.chrX 15357392 119 + 21770863 ACCGAAACCCAGGCCGAGACUUAAACUAAGCCCAAAACUGGACUUGG-ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUAGCGCCUGUCCACUGCGCUCCAAUUUCA .(((((..((((.(((((.((((...)))).(((....))).)))))-.))))..)))))..((((.((((.(((((......))))).))))((((..(....).))))))))...... ( -39.40, z-score = -2.70, R) >droEre2.scaffold_4690 7103424 119 + 18748788 ACCGAAACCCAGUCCGAGAUUAAGACUAAGCCCAAAACUGGACUUGG-ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACUGCGCUCCAAUUUCA .(((((..((((((((((.(((....)))..(((....))).)))))-)))))..)))))..((((..(((.(((((......))))).))).((((..(....).))))))))...... ( -41.00, z-score = -3.34, R) >consensus ACCGAAACCCAGUCCAAGACUAAGACUAAGCCCAAAACUGGACUGGG_ACUGGACUUCGGAAUGGACUAGGAAUGCGUUGUAUCGUGUGCCUUGCGCCUGUCCACGGCGCUCCAAUUUCA ...(((((((((((((......................)))))))))...((((...((...(((((.(((...(((..((((...))))..))).))))))))...)).)))).)))). (-30.89 = -31.21 + 0.32)


| Location | 16,745,746 – 16,745,865 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Shannon entropy | 0.09984 |
| G+C content | 0.52259 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -33.58 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16745746 119 - 22422827 UGAAAUUGGAGCGCCGUGGACAGGCGCAAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGUACCCAGUCCAGUUUUGGGCUUAGUCUUAGUCUUGGACUGGGUUUCGU- .(((((((((.((..((((((((((....)((............))...))).))))))..))...))))))(((((((((((..(((((......)))))..))))))))))))))..- ( -42.70, z-score = -2.50, R) >droSim1.chrX 12932480 120 - 17042790 UGAAAUUGGAGCGCCGUGGACAGGCGCAAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGUGCCCAGUCCAGUUUUGGGCUUAGUCUUAGUCUUGGACCGGGUUUCAGU ((((((((((.((..((((((((((....)((............))...))).))))))..))...))))))((((.((((((..(((((......)))))..)))))).)))))))).. ( -39.10, z-score = -1.20, R) >droSec1.super_17 573712 120 - 1527944 UGAAAUUGGAGCGCUGUGGACAGGCGCAAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGUGCCCAGUCCAGUUUUGGGCUUAGUCUUAGUCUUGGACCGGGUUUCAGU ((((((((((.((..((((((((((....)((............))...))).))))))..))...))))))((((.((((((..(((((......)))))..)))))).)))))))).. ( -40.00, z-score = -1.36, R) >droYak2.chrX 15357392 119 - 21770863 UGAAAUUGGAGCGCAGUGGACAGGCGCUAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGU-CCAAGUCCAGUUUUGGGCUUAGUUUAAGUCUCGGCCUGGGUUUCGGU ......((((((((.........))))((((....((......))....)))).))))..(((((..((((.-((((((((......))))))((.......)).)).))))..))))). ( -36.80, z-score = -0.84, R) >droEre2.scaffold_4690 7103424 119 - 18748788 UGAAAUUGGAGCGCAGUGGACAGGCGCAAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGU-CCAAGUCCAGUUUUGGGCUUAGUCUUAAUCUCGGACUGGGUUUCGGU ..............(((((((((((....)((............))...))).)))))))(((((..(((((-((((((((......))))))((.......)).)))))))..))))). ( -41.80, z-score = -2.63, R) >consensus UGAAAUUGGAGCGCAGUGGACAGGCGCAAGGCACACGAUACAACGCAUUCCUAGUCCAUUCCGAAGUCCAGU_CCCAGUCCAGUUUUGGGCUUAGUCUUAGUCUUGGACUGGGUUUCGGU ((((((((((.((..((((((((((....)((............))...))).))))))..))...))))))(((((((((((....((((...))))....)).))))))))))))).. (-33.58 = -34.50 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:42 2011