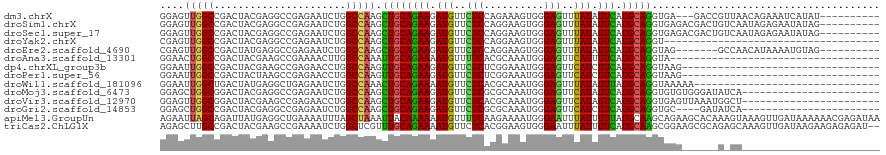
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,736,340 – 16,736,447 |
| Length | 107 |
| Max. P | 0.988305 |

| Location | 16,736,340 – 16,736,447 |
|---|---|
| Length | 107 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.59060 |
| G+C content | 0.47319 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -11.97 |
| Energy contribution | -11.54 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949657 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
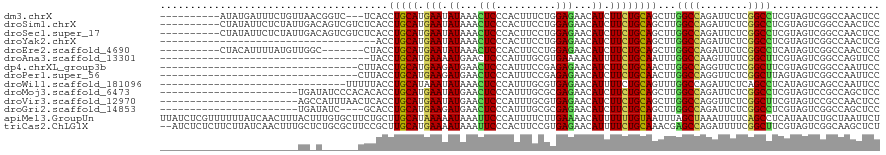
>dm3.chrX 16736340 107 + 22422827 ----------AUAUGAUUUCUGUUAACGGUC---UCACCUGCAUGAAUAUAAACUCCCACUUUCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCC ----------...........(((...((((---....(((((.(((.((...((((........))))...)).))))))))...))))........((((........)))))))... ( -26.30, z-score = -1.39, R) >droSim1.chrX 12925007 110 + 17042790 ----------CUAUAUUCUCUAUUGACAGUCGUCUCACCUGCAUGAAUAUAAACUCCCACUUCCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCC ----------..............(((....)))....(((((.(((.((...((((........))))...)).)))))))).((((((.((((..((....)).)))))))))).... ( -25.40, z-score = -1.24, R) >droSec1.super_17 564322 110 + 1527944 ----------CUAUAUUCUCUAUUGACAGUCGUCUCACCUGCAUGAAUAUAAACUCCCACUUCCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCC ----------..............(((....)))....(((((.(((.((...((((........))))...)).)))))))).((((((.((((..((....)).)))))))))).... ( -25.40, z-score = -1.24, R) >droYak2.chrX 15341404 84 + 21770863 ------------------------------------ACCUGCAUGAAUAUAAACUCCCACUUCCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCG ------------------------------------..(((((.(((.((...((((........))))...)).)))))))).((((((.((((..((....)).)))))))))).... ( -23.70, z-score = -2.31, R) >droEre2.scaffold_4690 7094162 103 + 18748788 ----------CUACAUUUUAUGUUGGC-------CUACCUGCAUGAAUAUAAACUCCCACUUCCUGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCAUAGUCGGCCAACUCG ----------...........((((((-------(.(((((((.(((.((...((((........))))...)).))))))))...((((........)))).....)).)))))))... ( -32.70, z-score = -4.20, R) >droAna3.scaffold_13301 1221 85 - 3000 -----------------------------------UACCUGCAUGAAAAUGAACUCCCAUUUGCGUGAAAACAUUUUCUGCAAUUUGGCCAAGUUUUCGGCUUCGUAGUCGGCCAGUUCC -----------------------------------....((((.(((((((...((.(......).))...)))))))))))..((((((..(....)((((....)))))))))).... ( -21.40, z-score = -1.55, R) >dp4.chrXL_group3b 38662 87 - 386183 ---------------------------------CUUACCUGCAUGAAGAUGAACUCCCAUUUCCGAGAGAACAUCUUCUGCAACUUGGCCAGGUUCUCGGCUUCGUAGUCGGCCAAUUCC ---------------------------------......((((.(((((((..(((.(......).)))..)))))))))))..((((((..(....)((((....)))))))))).... ( -27.60, z-score = -2.64, R) >droPer1.super_56 52618 87 - 431378 ---------------------------------CUUACCUGCAUGAAGAUGAACUCCCAUUUCCGAGAGAACAUCUUCUGCAACUUGGCCAGGUUCUCGGCUUAGUAGUCGGCCAAUUCC ---------------------------------......((((.(((((((..(((.(......).)))..)))))))))))..((((((..(....)((((....)))))))))).... ( -27.60, z-score = -2.68, R) >droWil1.scaffold_181096 4646675 89 - 12416693 -------------------------------UUUUUACCUGCAUAAAUAUAAACUCCCAUUUGCGUGAGAACAUUUUCUGCAGUUUGGCCAGAUUCUCAGCCUCAUAGUCAGCCAAUUCC -------------------------------.......(((((.(((......(((.(......).))).....))).))))).(((((..((((...........)))).))))).... ( -13.40, z-score = -0.44, R) >droMoj3.scaffold_6473 4392900 97 - 16943266 -----------------------UGAUAUCCCACACACCUGCAUGAAUAUGAACUCCCAUUUGCGCGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCCGCCAGCUCC -----------------------.(((...........(((((.(((.(((..(((.(......).)))..))).))))))))...((((........)))).....))).......... ( -19.70, z-score = 0.34, R) >droVir3.scaffold_12970 4849552 97 - 11907090 -----------------------AGCCAUUUAACUCACCUGCAUGAAUAUGAACUCCCAUUUGCGUGAGAACAUCUUCUGCAGCUUGGCCAGGUUCUCGGCUUCGUAGUCCGCCAACUCC -----------------------.((((..........(((((.(((.(((..(((.(......).)))..))).))))))))..))))..((((..(((((....)).)))..)))).. ( -21.30, z-score = -0.18, R) >droGri2.scaffold_14853 5515400 93 + 10151454 -----------------------UGAUAUC----GCACCUGCAUGAAGAUGAACUCCCAUUUGCGCGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAGCUCC -----------------------.......----((..(((((.(((((((..(((.(......).)))..))))))))))))..(((((.((((..((....)).)))))))))))... ( -32.00, z-score = -2.06, R) >apiMel3.GroupUn 86103497 120 + 399230636 UUAUCUCGUUUUUUAUCAACUUUACUUUGUGCUUCUGCUUGCAUAAAAAUAAAUUCCCAUUUUCUUGAAAACAUUUUUUGUAAUUUAGCUAAAUUUUCAGCCUCAUAAUCUGCUAAUUCU .......(((((.............(((((((........)))))))..........((......)))))))............(((((......................))))).... ( -10.65, z-score = -0.14, R) >triCas2.ChLG1X 8002018 118 - 8109244 --AUCUCUCUUCUUAUCAACUUUGCUCUGCGCUUCCGCUUGCAUGAAAAUAAAUUCCCACUUCCGUGAGAACAUUUUCUGCAAACGAGCCAGAUUUUCGGCUUCGUAGUCGGCAAGCUCU --..................(((((((((.(((..((.(((((.((((((...(((.(((....))).))).))))))))))).)))))))))....(((((....)))))))))).... ( -30.60, z-score = -2.55, R) >consensus _______________________UG_C______CUCACCUGCAUGAAUAUAAACUCCCAUUUCCGGGAGAACAUCUUCUGCAGCUUGGCCAGAUUCUCGGCCUCGUAGUCGGCCAACUCC ......................................(((((.(((.((...(((..........)))...)).))))))))...((((........)))).................. (-11.97 = -11.54 + -0.43)
| Location | 16,736,340 – 16,736,447 |
|---|---|
| Length | 107 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.59060 |
| G+C content | 0.47319 |
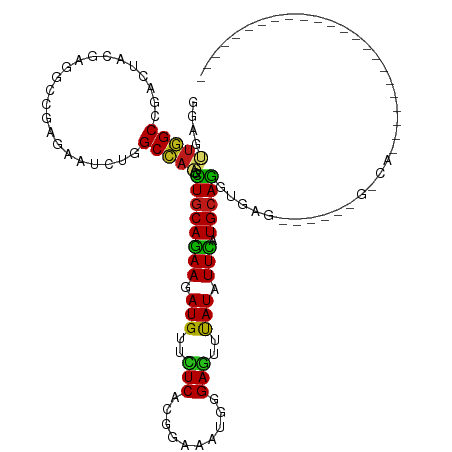
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -16.77 |
| Energy contribution | -16.09 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988305 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
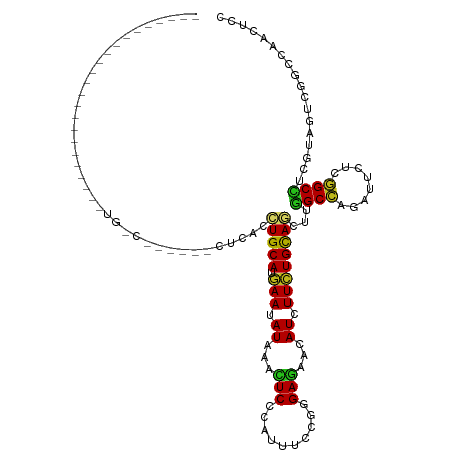
>dm3.chrX 16736340 107 - 22422827 GGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGAAAGUGGGAGUUUAUAUUCAUGCAGGUGA---GACCGUUAACAGAAAUCAUAU---------- ((..((((((........))))))...(((.(((....(((((((.(((..((((........))))..))).))).))))))).)---)))).................---------- ( -31.90, z-score = -1.70, R) >droSim1.chrX 12925007 110 - 17042790 GGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGGAAGUGGGAGUUUAUAUUCAUGCAGGUGAGACGACUGUCAAUAGAGAAUAUAG---------- ...((((((((.((((..(((((......)))))...((((((((.(((..((((........))))..))).))).))))))).)).))...))))))...........---------- ( -31.40, z-score = -1.00, R) >droSec1.super_17 564322 110 - 1527944 GGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGGAAGUGGGAGUUUAUAUUCAUGCAGGUGAGACGACUGUCAAUAGAGAAUAUAG---------- ...((((((((.((((..(((((......)))))...((((((((.(((..((((........))))..))).))).))))))).)).))...))))))...........---------- ( -31.40, z-score = -1.00, R) >droYak2.chrX 15341404 84 - 21770863 CGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGGAAGUGGGAGUUUAUAUUCAUGCAGGU------------------------------------ ((((((....)))).)).(((((......)))))...((((((((.(((..((((........))))..))).))).)))))..------------------------------------ ( -28.50, z-score = -1.76, R) >droEre2.scaffold_4690 7094162 103 - 18748788 CGAGUUGGCCGACUAUGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCCAGGAAGUGGGAGUUUAUAUUCAUGCAGGUAG-------GCCAACAUAAAAUGUAG---------- ...(((((((.((.....(((((......)))))...((((((((.(((..((((........))))..))).))).))))))).)-------))))))...........---------- ( -40.00, z-score = -4.37, R) >droAna3.scaffold_13301 1221 85 + 3000 GGAACUGGCCGACUACGAAGCCGAAAACUUGGCCAAAUUGCAGAAAAUGUUUUCACGCAAAUGGGAGUUCAUUUUCAUGCAGGUA----------------------------------- .....(((((((...((....)).....)))))))..((((((((((((..(((.((....)).)))..))))))).)))))...----------------------------------- ( -23.40, z-score = -1.48, R) >dp4.chrXL_group3b 38662 87 + 386183 GGAAUUGGCCGACUACGAAGCCGAGAACCUGGCCAAGUUGCAGAAGAUGUUCUCUCGGAAAUGGGAGUUCAUCUUCAUGCAGGUAAG--------------------------------- ....(((((((.((.((....))))....))))))).((((((((((((..((((((....))))))..))))))).))))).....--------------------------------- ( -33.30, z-score = -3.22, R) >droPer1.super_56 52618 87 + 431378 GGAAUUGGCCGACUACUAAGCCGAGAACCUGGCCAAGUUGCAGAAGAUGUUCUCUCGGAAAUGGGAGUUCAUCUUCAUGCAGGUAAG--------------------------------- ....(((((((....((......))....))))))).((((((((((((..((((((....))))))..))))))).))))).....--------------------------------- ( -32.60, z-score = -3.15, R) >droWil1.scaffold_181096 4646675 89 + 12416693 GGAAUUGGCUGACUAUGAGGCUGAGAAUCUGGCCAAACUGCAGAAAAUGUUCUCACGCAAAUGGGAGUUUAUAUUUAUGCAGGUAAAAA------------------------------- (((.(..(((........)))..)...))).......(((((..(((((((((((......)))))....)))))).))))).......------------------------------- ( -18.40, z-score = -0.15, R) >droMoj3.scaffold_6473 4392900 97 + 16943266 GGAGCUGGCGGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCGCGCAAAUGGGAGUUCAUAUUCAUGCAGGUGUGUGGGAUAUCA----------------------- (.((((..((.....)).(((((......))))).)))).)....(((((((.((((((..((.((((....))))...))..))))))))))))).----------------------- ( -34.90, z-score = -1.61, R) >droVir3.scaffold_12970 4849552 97 + 11907090 GGAGUUGGCGGACUACGAAGCCGAGAACCUGGCCAAGCUGCAGAAGAUGUUCUCACGCAAAUGGGAGUUCAUAUUCAUGCAGGUGAGUUAAAUGGCU----------------------- ((.(((..(((.((....)))))..)))))((((((.((((((((.(((..(((.((....)).)))..))).))).))))).)........)))))----------------------- ( -26.30, z-score = 0.07, R) >droGri2.scaffold_14853 5515400 93 - 10151454 GGAGCUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCGCGCAAAUGGGAGUUCAUCUUCAUGCAGGUGC----GAUAUCA----------------------- .((..(.(((........(((((......)))))...((((((((((((..(((.((....)).)))..))))))).)))))).))----.)..)).----------------------- ( -34.40, z-score = -1.34, R) >apiMel3.GroupUn 86103497 120 - 399230636 AGAAUUAGCAGAUUAUGAGGCUGAAAAUUUAGCUAAAUUACAAAAAAUGUUUUCAAGAAAAUGGGAAUUUAUUUUUAUGCAAGCAGAAGCACAAAGUAAAGUUGAUAAAAAACGAGAUAA ...((((((.........((((((....))))))...((((.(((((((((..((......))..)))..)))))).(((........)))....)))).)))))).............. ( -17.60, z-score = -0.52, R) >triCas2.ChLG1X 8002018 118 + 8109244 AGAGCUUGCCGACUACGAAGCCGAAAAUCUGGCUCGUUUGCAGAAAAUGUUCUCACGGAAGUGGGAAUUUAUUUUCAUGCAAGCGGAAGCGCAGAGCAAAGUUGAUAAGAAGAGAGAU-- ....((((.(((((.((....))....(((((((((((((((((((((((((((((....))))))))..)))))).))))))))..))).))))....))))).)))).........-- ( -42.80, z-score = -4.04, R) >consensus GGAGUUGGCCGACUACGAGGCCGAGAAUCUGGCCAAGCUGCAGAAGAUGUUCUCACGGAAAUGGGAGUUUAUAUUCAUGCAGGUGAG______G_CA_______________________ ....(((((......................))))).((((((((.(((..(((..........)))..))).))).)))))...................................... (-16.77 = -16.09 + -0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:38 2011