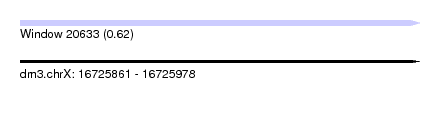
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,725,861 – 16,725,978 |
| Length | 117 |
| Max. P | 0.619803 |

| Location | 16,725,861 – 16,725,978 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 65.30 |
| Shannon entropy | 0.66678 |
| G+C content | 0.45448 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -11.43 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
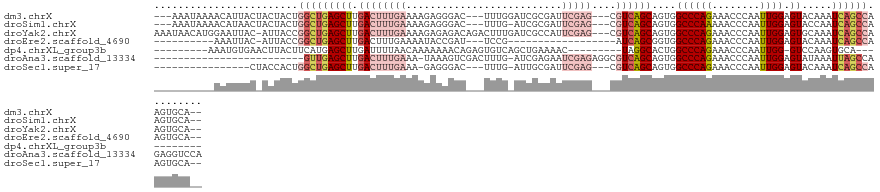
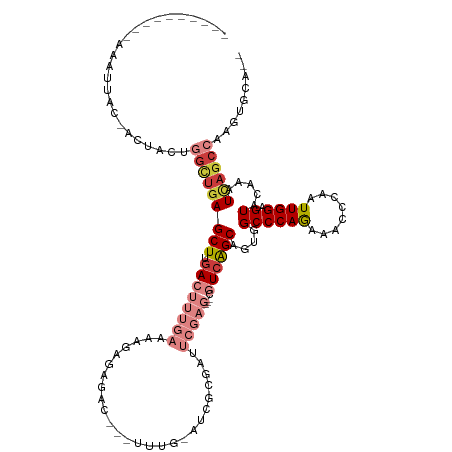
>dm3.chrX 16725861 117 + 22422827 ---AAAUAAAACAUUACUACUACUGGCUGAGCUUGACUUUGAAAAGAGGGAC---UUUGGAUCGCGAUUCGAG---CGUCAGCAGUGGCCCAGAAACCCAAUUGGAGUACAAAUCAGCCAAGUGCA-- ---.................((((((((((..(((((((..(.....(((..---(((((..(((.......(---(....)).)))..)))))..)))..)..)))).))).)))))).))))..-- ( -33.80, z-score = -1.94, R) >droSim1.chrX 12909522 116 + 17042790 ---AAAUAAAACAUAACUACUACUGGCUGAGCUUGACUUUGAAAAGAGGGAC---UUUG-AUCGCGAUUCGAG---CGUCAGCAGUGGCCCAAAAACCCAAUUGGAGUACCAAUCAGCCAAGUGCA-- ---.................((((((((((......((((....))))((((---((((-(.(((.......)---)))))).))).((((((........)))).)).))..)))))).))))..-- ( -29.70, z-score = -1.16, R) >droYak2.chrX 15331091 122 + 21770863 AAAUAACAUGGAAUUAC-AUUACCGGCUGAGCUUGACUUUGAAAAGAGAGACAGACUUUGAUCGCCAUUCGAG---CGUCAGCAGUGGCCCAGAAACCCAAUUGGAGUGCAAAUCAGCCAAGUGCA-- .......(((......)-))....((((((..(((((((..(....((((.....))))....((((((...(---(....))))))))............)..)))).))).)))))).......-- ( -27.70, z-score = -0.01, R) >droEre2.scaffold_4690 7083872 94 + 18748788 ----------AAAUUAC-AUUACCGGCUGAGCUUGACUUUGAAAAUACCGAU---UCCG------------------AUCAGCGGUGGCCCAGAAACCCAAUUGGAGUACAAAUCAGCCAAGUGCA-- ----------.......-......((((((..(((((((..(....((((..---....------------------.....))))((........))...)..)))).))).)))))).......-- ( -21.90, z-score = -1.01, R) >dp4.chrXL_group3b 13555 98 - 386183 ---------AAAUGUGAACUUACUUCAUGAGCUUGAUUUUAACAAAAAAACAGAGUGUCAGCUGAAAAC---------UAGGCACUGGCCCAGAAACCCAAUUGG-GUCCAAGUGCA----------- ---------....(((((.....)))))(.(((((..................((((((((.(....))---------).))))))(((((((........))))-)))))))).).----------- ( -22.20, z-score = -0.44, R) >droAna3.scaffold_13334 853696 101 - 1562580 -------------------------GUUGAGCUUGACUUUGAAA-UAAAGUCGACUUUG-AUCGAGAAUCGAGAGGCGUCAGCAGUGGCCCAGAAACCCAAUUGGAGUAUAAAUUAGCCAGAGGUCCA -------------------------((((((((((((((((...-))))))).......-.(((.....))).)))).)))))..((((......((((....)).))........))))........ ( -22.54, z-score = -0.18, R) >droSec1.super_17 554036 102 + 1527944 ----------------CUACCACUGGCUGAGCUUGACUUUGAAA-GAGGGAC---UUUG-AUUGCGAUUCGAG---CGUCAGCAGUGGCCCAGAAACCCAAUUGGAGUACAAAUCAGCCAAGUGCA-- ----------------....((((((((((..(((.((((....-)))).((---((..-((((.(.(((..(---.((((....)))))..))).).))))..)))).))).)))))).))))..-- ( -34.50, z-score = -2.29, R) >consensus __________AAAUUAC_ACUACUGGCUGAGCUUGACUUUGAAAAGAGAGAC___UUUG_AUCGCGAUUCGAG___CGUCAGCAGUGGCCCAGAAACCCAAUUGGAGUACAAAUCAGCCAAGUGCA__ ........................(((((((((.(((........................................))))))....((((((........)))).)).....))))))......... (-11.43 = -12.27 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:36 2011