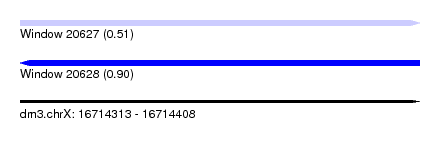
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,714,313 – 16,714,408 |
| Length | 95 |
| Max. P | 0.901157 |

| Location | 16,714,313 – 16,714,408 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 72.32 |
| Shannon entropy | 0.52393 |
| G+C content | 0.32448 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -9.39 |
| Energy contribution | -8.90 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
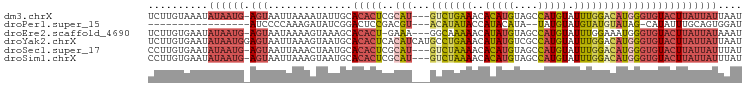
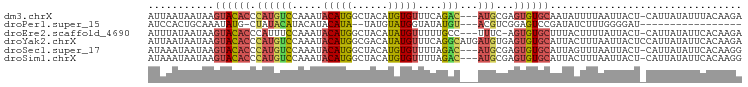
>dm3.chrX 16714313 95 + 22422827 UCUUGUAAAUAUAAUG-AGUAAUUAAAAUAUUGCACACUCGCAU---GUCUGAAACACAUGUAGCCAUGUAUUUGGACAUGGGUGUACUUAUUAUUAAU ..........((((((-((((((......)))))((((.(.(((---(((..((..(((((....))))).))..)))))))))))..))))))).... ( -24.60, z-score = -2.66, R) >droPer1.super_15 241039 76 + 2181545 -----------------AUCCCCAAAGAUAUCGGACUCCGACGU---ACAUAUACCAUACAUA--UAUGUAUGUAUGUAUAG-CAUAUUUGCAGUGGAU -----------------((((.(.......((((...)))).((---(((((...((((((..--..))))))))))))).(-((....))).).)))) ( -18.00, z-score = -1.29, R) >droEre2.scaffold_4690 7071162 94 + 18748788 UCUUGUGAAUAUAAUG-AGUAAUAAAAGUAAAGCACACU-GAAA---GGCAAAAACAUAUGUAGCCAUGUAUUUGGAAAUGGGUGUACUUAUUAUAAAU .........(((((((-(((.......(.....)(((((-.(..---..((((.((((........)))).))))....).)))))))))))))))... ( -16.00, z-score = -0.83, R) >droYak2.chrX 15319978 99 + 21770863 UCUUGUGAAUAUAAUGGAGUAAUUAAAGUAAUGCACACUCACAUCAUGCCUGAAACAUAUGUCGCCAUGUAUUUGGACAUGGGUGUACUUAUUAUUAAU .....................(((((((((((((((....((((.(((.......)))))))..((((((......))))))))))).))))).))))) ( -16.20, z-score = 1.15, R) >droSec1.super_17 542532 95 + 1527944 CCUUGUGAAUAUAAUG-AGUAAUUAAACUAAUGCACACUCGCAU---GUCUAAAACACAUGUAGCCAUGUAUUUGGACAUGGGUGUACUUAUUAUUUAU ..........((((((-(((.((((...))))..((((.(.(((---(((((((..(((((....))))).)))))))))))))))))))))))).... ( -24.20, z-score = -2.35, R) >droSim1.chrX 12894323 95 + 17042790 CCUUGUGAAUAUAAUG-AGUAAUUAAAGUAAUGCACACUCGCAU---GUCUAAAACACAUGUAGCCAUGUAUUUGGACAUGGGUGUACUUAUUAUUUAU ..........((((((-(((.......(.....)((((.(.(((---(((((((..(((((....))))).)))))))))))))))))))))))).... ( -25.10, z-score = -2.35, R) >consensus UCUUGUGAAUAUAAUG_AGUAAUUAAAGUAAUGCACACUCGCAU___GUCUAAAACACAUGUAGCCAUGUAUUUGGACAUGGGUGUACUUAUUAUUAAU ..................................(((((((......(((((((..(((((....))))).))))))).)))))))............. ( -9.39 = -8.90 + -0.49)
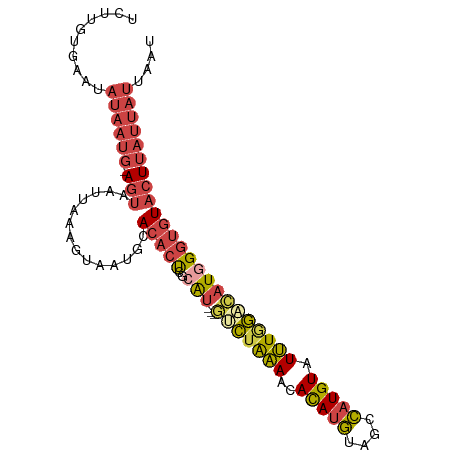
| Location | 16,714,313 – 16,714,408 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.32 |
| Shannon entropy | 0.52393 |
| G+C content | 0.32448 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -6.60 |
| Energy contribution | -7.55 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
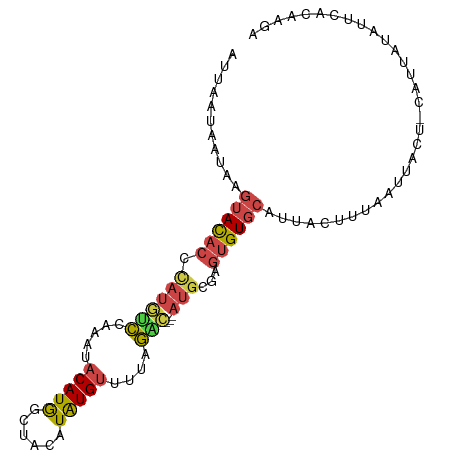
>dm3.chrX 16714313 95 - 22422827 AUUAAUAAUAAGUACACCCAUGUCCAAAUACAUGGCUACAUGUGUUUCAGAC---AUGCGAGUGUGCAAUAUUUUAAUUACU-CAUUAUAUUUACAAGA ...........(((((((((((((.(((((((((....)))))))))..)))---))).).))))))...............-................ ( -26.30, z-score = -4.24, R) >droPer1.super_15 241039 76 - 2181545 AUCCACUGCAAAUAUG-CUAUACAUACAUACAUA--UAUGUAUGGUAUAUGU---ACGUCGGAGUCCGAUAUCUUUGGGGAU----------------- .(((((((((..((((-(((((((((........--))))))))))))))))---).)).))).(((((.....)))))...----------------- ( -23.00, z-score = -2.22, R) >droEre2.scaffold_4690 7071162 94 - 18748788 AUUUAUAAUAAGUACACCCAUUUCCAAAUACAUGGCUACAUAUGUUUUUGCC---UUUC-AGUGUGCUUUACUUUUAUUACU-CAUUAUAUUCACAAGA .........((((((((.......((((.(((((......))))).))))..---....-.)))))))).............-................ ( -14.24, z-score = -2.03, R) >droYak2.chrX 15319978 99 - 21770863 AUUAAUAAUAAGUACACCCAUGUCCAAAUACAUGGCGACAUAUGUUUCAGGCAUGAUGUGAGUGUGCAUUACUUUAAUUACUCCAUUAUAUUCACAAGA ..................((((((.(((((.(((....))).)))))..)))))).((((((((((....................))))))))))... ( -19.45, z-score = -0.76, R) >droSec1.super_17 542532 95 - 1527944 AUAAAUAAUAAGUACACCCAUGUCCAAAUACAUGGCUACAUGUGUUUUAGAC---AUGCGAGUGUGCAUUAGUUUAAUUACU-CAUUAUAUUCACAAGG ....(((((..(((((((((((((.(((((((((....)))))))))..)))---))).).))))))...(((......)))-.))))).......... ( -26.30, z-score = -3.80, R) >droSim1.chrX 12894323 95 - 17042790 AUAAAUAAUAAGUACACCCAUGUCCAAAUACAUGGCUACAUGUGUUUUAGAC---AUGCGAGUGUGCAUUACUUUAAUUACU-CAUUAUAUUCACAAGG ...........(((((((((((((.(((((((((....)))))))))..)))---))).).))))))...............-................ ( -26.00, z-score = -3.94, R) >consensus AUUAAUAAUAAGUACACCCAUGUCCAAAUACAUGGCUACAUAUGUUUUAGAC___AUGCGAGUGUGCAUUACUUUAAUUACU_CAUUAUAUUCACAAGA ...........((((((((((((......))))))..((....))................))))))................................ ( -6.60 = -7.55 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:32 2011