
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,692,834 – 16,692,945 |
| Length | 111 |
| Max. P | 0.611800 |

| Location | 16,692,834 – 16,692,945 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.70869 |
| G+C content | 0.71217 |
| Mean single sequence MFE | -56.13 |
| Consensus MFE | -17.17 |
| Energy contribution | -16.77 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
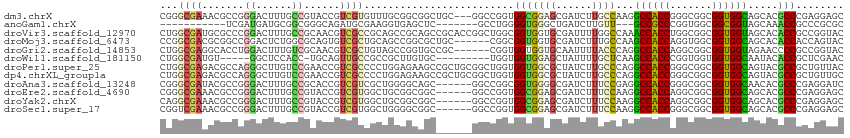
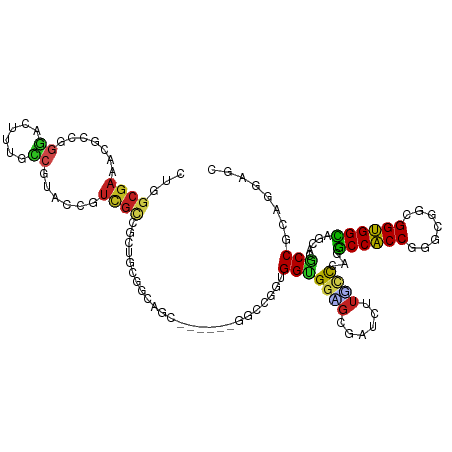
>dm3.chrX 16692834 111 + 22422827 CGGGCGAAACGCCGGGACUUUGCCGUACCGUCGUGUUUGCGGCGGCUGC---GGCCGGUGGCGGAGCGAUCUUGCCAAGGCCACCGGGCGGCGGUGGCAGCACGCCCGAGGAGC (((((....(((((.......(((((((((((((....))))))).)))---)))(((((((.(.(((....))))...)))))))..)))))(((....))))))))...... ( -60.10, z-score = -1.18, R) >anoGam1.chrX 16424552 93 - 22145176 -----------UCGAUGAUGCGGCGGGCAGAUGCGAAGGUGAGCUC-------GCCUGGGGUGGGCUGAUCUUGUU---GCCGCCCGGUGGCGGCGGUAGCAAACCGCCCGCGC -----------........(((.(((((.......(((((.(((((-------(((...)))))))).)))))(((---(((((((....).))))))))).....)))))))) ( -48.90, z-score = -1.98, R) >droVir3.scaffold_12970 4745656 114 - 11907090 CUGGCGAUGCGCCCGGACUUUGCCGCAACGUCGCCGCAGCCGCAGCCGCACCGGCUGGCGGUGGUGCGAUUUUGGCCAAACCACCUGGCGGCGGUGGUAGCACACCGCCGGUAC (.((((...)))).).....((((.(((.(((((.((.((((((((((...))))).))))).))))))).)))((((.......))))(((((((......))))))))))). ( -55.90, z-score = -0.54, R) >droMoj3.scaffold_6473 3321853 108 + 16943266 CCGGCGACGCGGCCGGACUCUGGCGCAGUGUCGCUGCAGCCGGCGCUGC------CGGCGGUGGUGCGAUCUUUGCCAAGCCACCAGGUGGCGGUGGCAGCACACCACCAGUAC .(((((((((.(((((...)))))...)))))))))..((((((...))------))))(((((((.(....((((((.(((((...)))))..))))))).)))))))..... ( -62.50, z-score = -2.59, R) >droGri2.scaffold_14853 1377696 108 - 10151454 CUGGCGAGGCACCUGGACUUUGUCGCAACGUCGCUGUAGCCGGUGCCGC------CGGUGGUGGUGCAAUUUUACCCAGGCCACCAGGCGGCGGUGGUAGAACCCCGCCGGUAC ..(((((.(....(((((...))).)).).)))))...(((((((((((------(((((((((((......))))...)))))).))))))((.((.....)))))))))).. ( -50.10, z-score = -0.89, R) >droWil1.scaffold_181150 3911953 99 + 4952429 CUGGCGAUGU-----GGCUCCACC-UGCAGUUGCCGCCGCUUGUGC---------UGGUGGUGGAGCUAUUUUGCUCAAGCCACCCGGUGGUGGUGGCAAUACACCGCUCGAAC (.((((.(((-----.((......-.)).((((((((((((...((---------(((((((.((((......))))..)))).)))))))))))))))))))).)))).)... ( -46.30, z-score = -2.57, R) >droPer1.super_25 784284 114 - 1448063 CUGGCGAGACGCCAGGGCUUGUCCGAACCGUCGCCCCUGGAGAAGCCGCUGCGGCUGGUGGUGGCGCUAUCUUGCCCAGGCCACCGGGCGGCGGUGGCAGUACGCCGCUGUUAC ((((((...))))))(((........((((((((((.......(((((...)))))((((((((.((......))))..))))))))))))))))(((.....))))))..... ( -58.10, z-score = -1.08, R) >dp4.chrXL_group1a 6647452 114 + 9151740 CUGGCGAGACGCCAGGGCUUGUCCGAACCGUCGCCCCUGGAGAAGCCGCUGCGGCUGGUGGUGGCGCUAUCUUGCCCAGGCCACCGGGCGGCGGUGGCAGUACGCCGCUGUUGC ((((((...))))))(((........((((((((((.......(((((...)))))((((((((.((......))))..))))))))))))))))(((.....))))))..... ( -58.10, z-score = -0.69, R) >droAna3.scaffold_13248 525701 108 - 4840945 CGGGCGAUACGCCGGGACUUUGCCGCACCGUCGUCGCUGGGGCAGC------GGCCGGCGGUGGGGCGAUCUUUCCGAGGCCACCGGGCGGCGGUGGCAACACGCCCGAGGAUC ((((((......(((((..(((((.((((((((((((((...))))------)).)))))))).)))))...)))))..(((((((.....)))))))....))))))...... ( -66.00, z-score = -3.67, R) >droEre2.scaffold_4690 7049591 108 + 18748788 CGGGCGAAACGCCGGGACUUUGCCGUACCGUCGUGGCUGCGGCGGC------GGCCGGUGGCGGAGCGAUCUUUCCAAGGCCACCAGGCGGCGGUGGCAGCACGCCCGAGGAGC ((((((..(((.(((.((......)).))).))).(((((.((.((------.(((((((((((((......))))...)))))).))).)).)).))))).))))))...... ( -60.30, z-score = -1.98, R) >droYak2.chrX 15299157 108 + 21770863 CAGGCGAAACGCCGGGACUUUGCCGUACCGUCGUGGCUGCGGCGGC------GGCCGGUGGCGGAGCGAUCUUUCCAAGGCCACCAGGCGGCGGUGGCAGCACGCCCGAGGAGC ..((((...))))(((...(((((..((((((((((((((....))------))))((((((((((......))))...))))))..)))))))))))))....)))....... ( -56.00, z-score = -1.16, R) >droSec1.super_17 521862 108 + 1527944 CGGUCGAAACGCCGGGACUUUGCCGUACCGUCGUGGCUGGGGCGGC------GGCCGGUGGCGGAGCGAUCUUUCCAAGGCCACCGGGCGGCGGUGGCAGCACGCCCGAGGAGC .(((......)))(((...(((((..((((((((.((....)).))------(.((((((((((((......))))...)))))))).))))))))))))....)))....... ( -51.30, z-score = 0.46, R) >consensus CUGGCGAAACGCCGGGACUUUGCCGUACCGUCGCCGCUGCGGCAGC______GGCCGGUGGUGGAGCGAUCUUGCCCAGGCCACCGGGCGGCGGUGGCAGCACGCCGCAGGAGC ...((((.......((......))......)))).........................(((((((......))))...((((((.......)))))).....)))........ (-17.17 = -16.77 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:28 2011