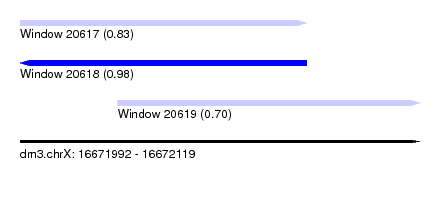
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,671,992 – 16,672,119 |
| Length | 127 |
| Max. P | 0.980427 |

| Location | 16,671,992 – 16,672,083 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.85 |
| Shannon entropy | 0.55230 |
| G+C content | 0.49565 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.78 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
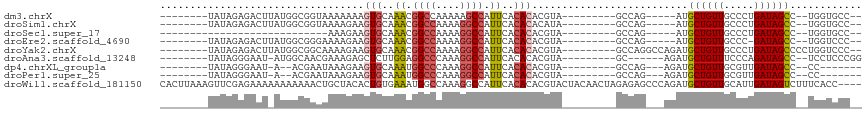
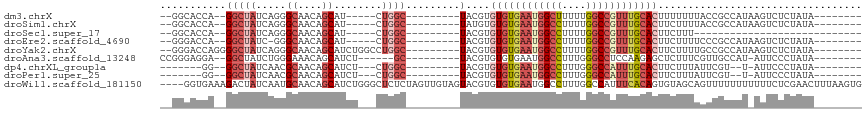
>dm3.chrX 16671992 91 + 22422827 --------UAUAGAGACUUAUGGCGGUAAAAAAAGUGCAAACGGCCAAAAAGCCAUUCACACACGUA---------GCCAG-----AUGCUGUUGCCCUGAUAGCC--UGGUGCC-- --------.............(((..........(((..((.(((......))).))..))).....---------(((((-----..((((((.....)))))))--)))))))-- ( -22.70, z-score = -0.36, R) >droSim1.chrX 12851053 91 + 17042790 --------UAUAGAGACUUAUGGCGGUAAAAGAAGUGCAAACGGCCAAAAGGCCAUUCACACACAUA---------GCCAG-----AUGCUGUUGCCCUGAUAGCC--UGGUGCC-- --------.............(((..........(((..((.((((....)))).))..))).....---------(((((-----..((((((.....)))))))--)))))))-- ( -26.30, z-score = -1.10, R) >droSec1.super_17 500679 71 + 1527944 ----------------------------AAAGAAGUGCAAACGGCCAAAAGGCCAUUCACACACGUA---------GCCAG-----AUGCUGUUGCCCUGAUAGCC--UGGUGCC-- ----------------------------......(((..((.((((....)))).))..))).....---------(((((-----..((((((.....)))))))--))))...-- ( -22.50, z-score = -1.60, R) >droEre2.scaffold_4690 7029482 90 + 18748788 --------UAUAGAGACUUAUGGCGGGAAAAGAAGUGCAAACGGCCAAAAGGCCAUUCACACACGUA---------GCCAG-----AUGCUGUUGCCC-GAUAGCC--UGGUCCC-- --------................((((......(((..((.((((....)))).))..))).....---------.((((-----..((((((....-)))))))--)))))))-- ( -26.00, z-score = -1.09, R) >droYak2.chrX 15278926 98 + 21770863 --------UAUAGAGACUUAUGGCGGCAAAAGAAGUGCAAACGGCCAAAAGGCCAUUCACACACGUA---------GCCAGGCCAGAUGCUGUUGCCCUGAUAGCCCCUGGUCCC-- --------............((((.(((.......)))....((((....)))).............---------))))((((((..((((((.....))))))..))))))..-- ( -31.90, z-score = -1.67, R) >droAna3.scaffold_13248 4299958 91 + 4840945 --------UAUAGGGAAU-AUGGCAACGAAAGAGCUCUUGGAGGCCCAAAGGCCAUUCACACACGUA---------GC------AGAUGCUGUUUCCCAGAUAGCC--UCCUCCCGG --------....((((..-..(((..(....)...(((.(((((((....))))...........((---------((------....))))..))).)))..)))--...)))).. ( -28.90, z-score = -1.66, R) >dp4.chrXL_group1a 2486198 85 - 9151740 --------UAUAGGGAAU-A--ACGAAUAAAGAAGUGCAAAUGGCCCAAAGGCCAUUCACACACGUA---------GCCAG---AGAUGCUGUUGCGUUGAUAGCC--CC------- --------....(((...-.--............(((..(((((((....)))))))..)))(((((---------((.((---.....)))))))))......))--).------- ( -25.80, z-score = -3.24, R) >droPer1.super_25 645502 85 + 1448063 --------UAUAGGGAAU-A--ACGAAUAAAGAAGUGCAAAUGGCCCAAAGGCCAUUCACACACGUA---------GCCAG---AGAUGCUGUUGCGUUGAUAGCC--CC------- --------....(((...-.--............(((..(((((((....)))))))..)))(((((---------((.((---.....)))))))))......))--).------- ( -25.80, z-score = -3.24, R) >droWil1.scaffold_181150 927066 113 - 4952429 CACUUAAAGUUCGAGAAAAAAAAAAACUGCUACACUGUGAAAUGGCCAAAGGCCAUUCACACACGUACUACAACUAGAGAGCCCAGAUGCUGUUGCAUUGAUAGUCUUUCACC---- ..............................(((..((((((.(((((...)))))))))))...))).........((((((...(((((....)))))....).)))))...---- ( -23.90, z-score = -1.14, R) >consensus ________UAUAGAGAAU_AUGGCGACAAAAGAAGUGCAAACGGCCAAAAGGCCAUUCACACACGUA_________GCCAG___AGAUGCUGUUGCCCUGAUAGCC__UGGUCCC__ ..................................(((..((.((((....)))).))..)))..........................((((((.....))))))............ (-11.40 = -11.78 + 0.37)
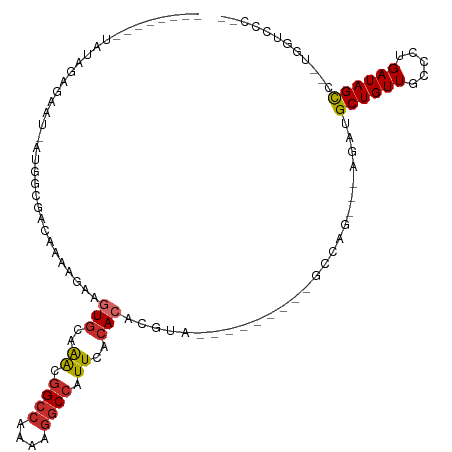
| Location | 16,671,992 – 16,672,083 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.85 |
| Shannon entropy | 0.55230 |
| G+C content | 0.49565 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16671992 91 - 22422827 --GGCACCA--GGCUAUCAGGGCAACAGCAU-----CUGGC---------UACGUGUGUGAAUGGCUUUUUGGCCGUUUGCACUUUUUUUACCGCCAUAAGUCUCUAUA-------- --(((....--(((...((((((....)).)-----)))))---------)....(((..(((((((....)))))))..)))..........))).............-------- ( -29.00, z-score = -1.66, R) >droSim1.chrX 12851053 91 - 17042790 --GGCACCA--GGCUAUCAGGGCAACAGCAU-----CUGGC---------UAUGUGUGUGAAUGGCCUUUUGGCCGUUUGCACUUCUUUUACCGCCAUAAGUCUCUAUA-------- --(((....--(((...((((((....)).)-----)))))---------)....(((..(((((((....)))))))..)))..........))).............-------- ( -31.40, z-score = -1.97, R) >droSec1.super_17 500679 71 - 1527944 --GGCACCA--GGCUAUCAGGGCAACAGCAU-----CUGGC---------UACGUGUGUGAAUGGCCUUUUGGCCGUUUGCACUUCUUU---------------------------- --(((....--.))).(((((((....)).)-----)))).---------.....(((..(((((((....)))))))..)))......---------------------------- ( -26.10, z-score = -1.75, R) >droEre2.scaffold_4690 7029482 90 - 18748788 --GGGACCA--GGCUAUC-GGGCAACAGCAU-----CUGGC---------UACGUGUGUGAAUGGCCUUUUGGCCGUUUGCACUUCUUUUCCCGCCAUAAGUCUCUAUA-------- --(((((..--(((....-(((....(((..-----...))---------)....(((..(((((((....)))))))..))).......))))))....)))))....-------- ( -33.30, z-score = -2.63, R) >droYak2.chrX 15278926 98 - 21770863 --GGGACCAGGGGCUAUCAGGGCAACAGCAUCUGGCCUGGC---------UACGUGUGUGAAUGGCCUUUUGGCCGUUUGCACUUCUUUUGCCGCCAUAAGUCUCUAUA-------- --(((((....(((..(((((((....)).)))))...(((---------.....(((..(((((((....)))))))..))).......))))))....)))))....-------- ( -41.30, z-score = -2.77, R) >droAna3.scaffold_13248 4299958 91 - 4840945 CCGGGAGGA--GGCUAUCUGGGAAACAGCAUCU------GC---------UACGUGUGUGAAUGGCCUUUGGGCCUCCAAGAGCUCUUUCGUUGCCAU-AUUCCCUAUA-------- ..(((((((--((((....(((....(((....------))---------).(((......))).)))...)))))))..(.((.........)))..-..))))....-------- ( -27.90, z-score = 0.03, R) >dp4.chrXL_group1a 2486198 85 + 9151740 -------GG--GGCUAUCAACGCAACAGCAUCU---CUGGC---------UACGUGUGUGAAUGGCCUUUGGGCCAUUUGCACUUCUUUAUUCGU--U-AUUCCCUAUA-------- -------((--(((((.....((....))....---.))))---------.....(((..(((((((....)))))))..)))............--.-...)))....-------- ( -28.10, z-score = -3.17, R) >droPer1.super_25 645502 85 - 1448063 -------GG--GGCUAUCAACGCAACAGCAUCU---CUGGC---------UACGUGUGUGAAUGGCCUUUGGGCCAUUUGCACUUCUUUAUUCGU--U-AUUCCCUAUA-------- -------((--(((((.....((....))....---.))))---------.....(((..(((((((....)))))))..)))............--.-...)))....-------- ( -28.10, z-score = -3.17, R) >droWil1.scaffold_181150 927066 113 + 4952429 ----GGUGAAAGACUAUCAAUGCAACAGCAUCUGGGCUCUCUAGUUGUAGUACGUGUGUGAAUGGCCUUUGGCCAUUUCACAGUGUAGCAGUUUUUUUUUUUCUCGAACUUUAAGUG ----.(.((((((.......(((....(((.((((.....)))).)))..((((((((.((((((((...)))))))))))).))))))).......)))))).)............ ( -29.44, z-score = -0.92, R) >consensus __GGGACCA__GGCUAUCAGGGCAACAGCAUCU___CUGGC_________UACGUGUGUGAAUGGCCUUUUGGCCGUUUGCACUUCUUUUGCCGCCAU_AGUCUCUAUA________ .....................((....))..........................((((((((((((....)))))))))))).................................. (-15.65 = -16.39 + 0.74)
| Location | 16,672,023 – 16,672,119 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Shannon entropy | 0.46107 |
| G+C content | 0.55868 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
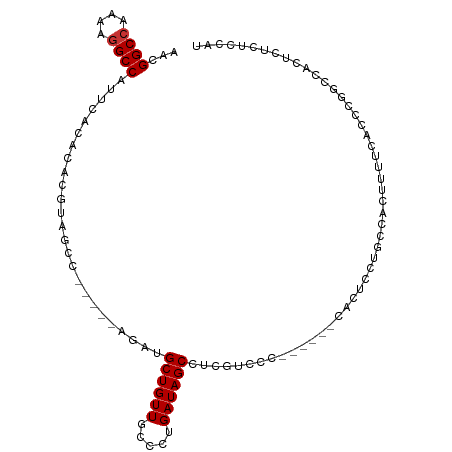
>dm3.chrX 16672023 96 + 22422827 AACGGCCAAAAAGCCAUUCACACACGUAGCC-----AGAUGCUGUUGCCCUGAUAGCCUGGUGCC------CACUCCUUCCACUUUUCACCCGGCCACUCUCUCCAU ...((((............((....)).(((-----((..((((((.....)))))))))))...------.....................))))........... ( -20.00, z-score = -0.83, R) >droSim1.chrX 12851084 96 + 17042790 AACGGCCAAAAGGCCAUUCACACACAUAGCC-----AGAUGCUGUUGCCCUGAUAGCCUGGUGCC------CACUUCCUCCACUUUUCACCCGGCCACUCUCUCCAU ...((((...((((...(((..((.(((((.-----....)))))))...)))..))))((((..------................)))).))))........... ( -21.97, z-score = -1.12, R) >droSec1.super_17 500690 96 + 1527944 AACGGCCAAAAGGCCAUUCACACACGUAGCC-----AGAUGCUGUUGCCCUGAUAGCCUGGUGCC------CACUACUGCCACUUUUCACCCGGCCACUCUCUCCAU ...((((....)))).............(((-----((..((((((.....)))))))))))...------.......(((...........)))............ ( -22.30, z-score = -0.27, R) >droEre2.scaffold_4690 7029513 95 + 18748788 AACGGCCAAAAGGCCAUUCACACACGUAGCC-----AGAUGCUGUUGCCC-GAUAGCCUGGUCCC------CACUCUUUCCACUUUUCACCCGGCCACUCUCUCCAU ...((((....(........)....(..(((-----((..((((((....-)))))))))))..)------.....................))))........... ( -20.80, z-score = -1.16, R) >droYak2.chrX 15278957 97 + 21770863 AACGGCCAAAAGGCCAUUCACACACGUAGCCAGGCCAGAUGCUGUUGCCCUGAUAGCCCCUGGUC----------CCUGGUUCAUCCACUCCUUCCACUCUCUCCAU ...((((....))))..........(.(((((((((((..((((((.....))))))..))))..----------))))))))........................ ( -30.30, z-score = -2.93, R) >droAna3.scaffold_13248 4299988 101 + 4840945 GGAGGCCCAAAGGCCAUUCACACACGUAGC------AGAUGCUGUUUCCCAGAUAGCCUCCUCCCGGAAAGCACUUUCCACCCAUCCAUUCCGUCCACUCUCUCUAU (((((((....)))).........((....------.((.((((((.....)))))).)).....(((((....)))))............)))))........... ( -22.00, z-score = -1.54, R) >dp4.chrXL_group1a 2486226 90 - 9151740 AAUGGCCCAAAGGCCAUUCACACACGUAGCCAG---AGAUGCUGUUGCGUUGAUAGCCCCGACUC-------------GGCCCGAGU-UCCAGCACUCUCUCCCUAU (((((((....))))))).......((((..((---((((((((..((.(((...(((.......-------------))).)))))-..))))).)))))..)))) ( -30.30, z-score = -3.12, R) >droPer1.super_25 645530 90 + 1448063 AAUGGCCCAAAGGCCAUUCACACACGUAGCCAG---AGAUGCUGUUGCGUUGAUAGCCCCGACUC-------------GGCCCGAGU-UCCAGCACUCUCUCCCUAU (((((((....))))))).......((((..((---((((((((..((.(((...(((.......-------------))).)))))-..))))).)))))..)))) ( -30.30, z-score = -3.12, R) >consensus AACGGCCAAAAGGCCAUUCACACACGUAGCC_____AGAUGCUGUUGCCCUGAUAGCCUCGUCCC______CACUCCUGCCACUUUUCACCCGGCCACUCUCUCCAU ...((((....)))).........................((((((.....)))))).................................................. (-11.90 = -12.03 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:24 2011