| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,662,725 – 16,662,787 |
| Length | 62 |
| Max. P | 0.903799 |

| Location | 16,662,725 – 16,662,787 |
|---|---|
| Length | 62 |
| Sequences | 9 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.39499 |
| G+C content | 0.52079 |
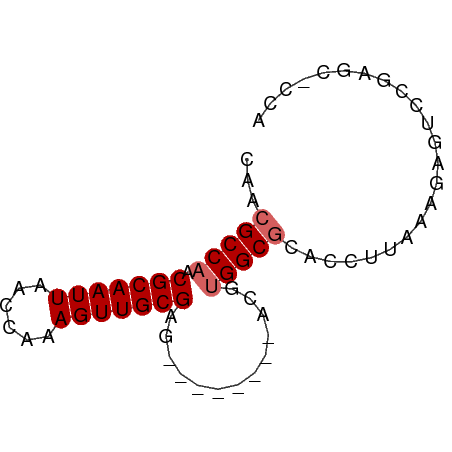
| Mean single sequence MFE | -14.89 |
| Consensus MFE | -10.37 |
| Energy contribution | -10.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903799 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 16662725 62 + 22422827 CAACGCCAACGCAAUUAACAAAAGUUGCGAG--------ACGUGGCGCACCUUAAAGAGUCCGAGCACCA ...(((((.(((((((......)))))))..--------...)))))....................... ( -14.40, z-score = -1.26, R) >droSec1.super_17 491303 62 + 1527944 CAACGCCAACGCAAUUAACAAAAGUUGCGAG--------ACGUGGCGCACCUUAAAGAGUCCGAGCACCA ...(((((.(((((((......)))))))..--------...)))))....................... ( -14.40, z-score = -1.26, R) >droYak2.chrX 15269576 62 + 21770863 CAACGCCAACGCAAUUAACAAAAGUUGCGAG--------ACGUGGCGCACCUUUAAGAGUCCGAGUUCCA ...(((((.(((((((......)))))))..--------...)))))....................... ( -14.40, z-score = -1.08, R) >droEre2.scaffold_4690 7020323 62 + 18748788 CAACGCCAACGCAAUUAACAAAAGUUGCGAG--------ACGUGGCACACCUUAAAGAGUCCGAGUUCCA ....((((.(((((((......)))))))..--------...))))........................ ( -13.10, z-score = -1.25, R) >droAna3.scaffold_13248 4284579 67 + 4840945 CAACGCCAACGCAAUUAACCAAAGUUGCGAGUCUCCAGGCCAGGGCGCACCUUCAAGA---CUCGUCCCA ...((((..(((((((......))))))).(((....)))...))))...........---......... ( -15.70, z-score = -0.99, R) >dp4.chrXL_group1a 2475673 56 - 9151740 CAACGCCAACGCAAUUAACCAAAGUUGCGAG--------ACAUGGCACACCUUAAAGAGCCCCC------ ....((((.(((((((......)))))))..--------...))))..................------ ( -12.00, z-score = -2.93, R) >droVir3.scaffold_12970 6389435 57 + 11907090 CAACGCCAACGCAAUUAACCAAAGUUGCGA---------CUGGGGCGCACCUUAAACAGCGCACGC---- .....(((.(((((((......))))))).---------.))).((((..........))))....---- ( -15.70, z-score = -1.03, R) >droMoj3.scaffold_6473 1748164 61 - 16943266 CAACGCCAACGCAAUUAACCAAAGUUGCGA---------CUGUGGCGCACCUUAAAAGGCGCUCGCUUCA ....((((.(((((((......))))))).---------...))))((.(((....))).))........ ( -19.20, z-score = -2.05, R) >droGri2.scaffold_15203 2648599 50 - 11997470 CAACGCCAACGCAAUUAACCAAAGUUGCGA---------CUGUGGCGCACCUUAAACAG----------- ...(((((.(((((((......))))))).---------...)))))............----------- ( -15.10, z-score = -3.07, R) >consensus CAACGCCAACGCAAUUAACCAAAGUUGCGAG________ACGUGGCGCACCUUAAAGAGUCCGAGC_CCA ...(((((.(((((((......))))))).............)))))....................... (-10.37 = -10.82 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:22 2011