
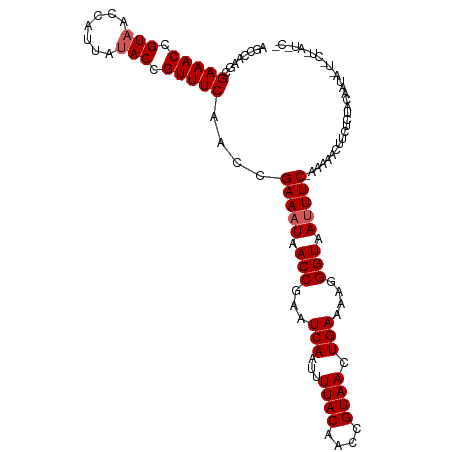
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,617,041 – 16,617,283 |
| Length | 242 |
| Max. P | 0.998319 |

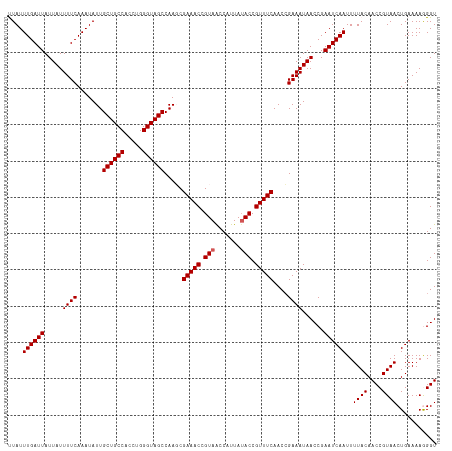
| Location | 16,617,041 – 16,617,142 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.24696 |
| G+C content | 0.36277 |
| Mean single sequence MFE | -16.84 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16617041 101 - 22422827 AGCCAAGCGAAACCGUAGCCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAAUUC-UAAAACUUCUCUACCAU------------ ........(((((.(((.......))).)))))....(((.(.(((.........((((....))))((....))))).).)))-.................------------ ( -12.40, z-score = -0.47, R) >droEre2.scaffold_4690 6974887 112 - 18748788 AGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACGGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGUUAUUUC-AAAAACAUCUCUACUAUAAUGCUCAUGC- (((.....(((((.(((.......))).)))))....(((((((((...(((...((((....)))).)))....)))))))))-....................))).....- ( -20.30, z-score = -3.14, R) >droYak2.chrX 15222440 99 - 21770863 AGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGUAAUUUCGAGAAACAUUUCUAC--------------- ........(((((.(((.......))).)))))...((((((.(((...(((...((((....)))).)))....))).))))))((((....))))..--------------- ( -17.90, z-score = -3.12, R) >droSec1.super_17 440838 113 - 1527944 AGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUC-GAAAACUUCCCUACAAUAUUACUAAUACG ........(((((.(((.......))).)))))...((((((.(((.........((((....))))((....))))).)))))-)............................ ( -17.60, z-score = -2.10, R) >droSim1.chrX 12762962 113 - 17042790 AGCCAAGCGAAACCGUAACCAUUAAACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUC-GAAAACUUCUCUACAAUAUUACUAAUACG ........(((((.((.........)).)))))...((((((.(((.........((((....))))((....))))).)))))-)............................ ( -16.00, z-score = -1.37, R) >consensus AGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGUAAUUUC_AAAAACUUCUCUACAAUA_U_CU_AU_C_ ........(((((.(((.......))).)))))....(((((.(((...(((...((((....)))).)))....))).))))).............................. (-12.36 = -12.76 + 0.40)


| Location | 16,617,064 – 16,617,182 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.46 |
| Shannon entropy | 0.04304 |
| G+C content | 0.36610 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16617064 118 - 22422827 UUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAGCCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGU ....((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))..((((....))))((....))... ( -23.90, z-score = -1.39, R) >droEre2.scaffold_4690 6974921 118 - 18748788 CUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACGGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGU ....((((((.((((((((.......((((((.....)))))).....(((((.(((.......))).)))))...))))))))...)))))).......(((............))) ( -25.60, z-score = -2.51, R) >droYak2.chrX 15222461 118 - 21770863 CUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAACCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAAGGU .(((((((.........)))))))..((((((.....))))))..((((((((.(((.......))).)))))................(((...((((....)))).)))....))) ( -22.30, z-score = -1.97, R) >droSec1.super_17 440873 118 - 1527944 UUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGU ....((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))..((((....))))((....))... ( -23.10, z-score = -1.41, R) >droSim1.chrX 12762997 118 - 17042790 UUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAAACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGU ....((((((.....((((.......((((((.....)))))).....(((((.((.........)).)))))....))))......))))))..((((....))))((....))... ( -21.50, z-score = -0.87, R) >consensus UUAUUUGAUUAUUAUUUUCAAAUAUUGCUGCCACCUGGGUAGCCAAGCGAAACCGUAACCAUUAUACCGUUUCAACCGAAAUAACCGAAUCAAUUUUACAACCGUAACUGAAAAGGGU ....((((((.....((((.......((((((.....)))))).....(((((.(((.......))).)))))....))))......))))))..((((....))))........... (-21.48 = -21.68 + 0.20)


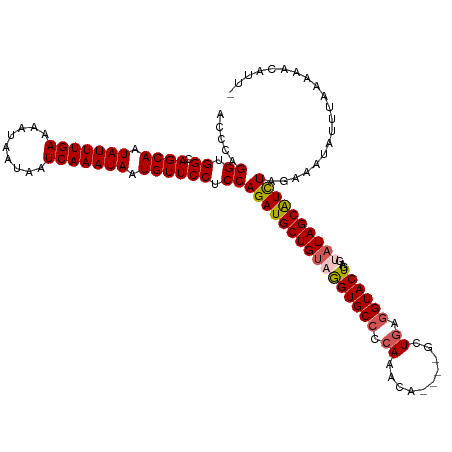
| Location | 16,617,142 – 16,617,257 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Shannon entropy | 0.15334 |
| G+C content | 0.36213 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16617142 115 + 22422827 ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAACA----GCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAACUUU- .....((.((.((((.(((((((.........))))))).)))))).))((((((((((((((((.((....----..)).))))))...))))))))))...................- ( -34.70, z-score = -4.22, R) >droEre2.scaffold_4690 6974999 119 + 18748788 ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGAAGGUGCCUCAUACAUAUAGCUGAAGUACUGAGAAUAGCGUCUACAAUUAUUUUAAAACAUU- .....((.((.((((.(((((((.........))))))).)))))).))((((((((..(((((.(((..........))).))))).....))))))))...................- ( -28.80, z-score = -2.31, R) >droYak2.chrX 15222539 115 + 21770863 ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAGUGUUCCUCCAGAUGCUGUAGGUGCCCCAAACA----GCUGAGGUACUAAGAAUAGCAUUUACAAUUAUUUAAAAGCAUU- .....((.((.((((.(((((((.........))))))).)))))).))(((((((((.((((((.((....----..)).))))))....)))))))))...................- ( -30.30, z-score = -2.77, R) >droSec1.super_17 440951 115 + 1527944 ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAACA----GCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAACUUU- .....((.((.((((.(((((((.........))))))).)))))).))((((((((((((((((.((....----..)).))))))...))))))))))...................- ( -34.70, z-score = -4.22, R) >droSim1.chrX 12763075 116 + 17042790 ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAAGUGCCCCAAACA----GCUCAGGUACUGAGUAUAGCAUCUAGAAACAUUUAAAAAACUUU .....((.((.((((.(((((((.........))))))).)))))).))((((((((((((((((.......----.....))))))...)))))))))).................... ( -32.10, z-score = -4.22, R) >consensus ACCCAGGUGGCAGCAAUAUUUGAAAAUAAUAAUCAAAUAAUGUUCCUCCAGAUGCUGUAGGUGCCCCAAACA____GCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAACAUU_ .....((.((.((((.(((((((.........))))))).)))))).))((((((((((((((((.((..........)).))))))...)))))))))).................... (-30.24 = -30.76 + 0.52)


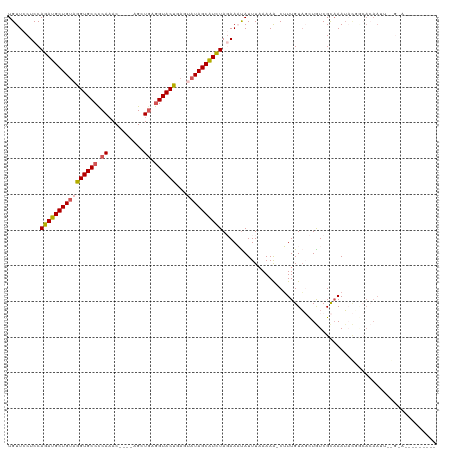
| Location | 16,617,182 – 16,617,283 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.43117 |
| G+C content | 0.39359 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16617182 101 + 22422827 UGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAA-CUUUGUAGCUGUCGCAAAUACGCCACAAUU-------------- ......((.((((((((((((((((.((...----...)).))))))...)))))))))).))............-.........(((.((......)).)))...-------------- ( -25.50, z-score = -1.81, R) >droEre2.scaffold_4690 6975039 95 + 18748788 UGUUCCUCCAGAUGCUGAAGGUGCCUCAUACAUAUAGCUGAAGUACUGAGAAUAGCGUCUACAAUUAUUUUAAAA-CAUUGUACUUAUCGCCAAUU------------------------ .........((((((((..(((((.(((..........))).))))).....))))))))(((((..........-.)))))..............------------------------ ( -15.50, z-score = 0.07, R) >droYak2.chrX 15222579 108 + 21770863 UGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUAAGAAUAGCAUUUACAAUUAUUUAAAAG-CAUUGUUCUUAUCGUAAAGUUGAAAAAAAGUAGCAGU------- .........(((((((((.((((((.((...----...)).))))))....)))))))))...............-.(((((((((.(((......)))....))).))))))------- ( -20.20, z-score = -0.44, R) >droSec1.super_17 440991 115 + 1527944 UGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC----AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAA-CUUUGUAGCUGUCGCAAAUACUGCACAAUUGGGAAGGUAGAGAU .....(((.((((((((((((((((.((...----...)).))))))...))))))))))...............-((((.(((.(((.(((.....)))))).))).))))...))).. ( -32.90, z-score = -1.83, R) >droSim1.chrX 12763115 116 + 17042790 UGUUCCUCCAGAUGCUGUAAGUGCCCCAAAC----AGCUCAGGUACUGAGUAUAGCAUCUAGAAACAUUUAAAAAACUUUGUAGCUGUCGCAAAUACUGCACAAUUGGGAAAGUAGAGAU ((((..((.((((((((((((((((......----......))))))...)))))))))).))))))........(((((.(...(((.(((.....))))))....).)))))...... ( -27.90, z-score = -1.01, R) >consensus UGUUCCUCCAGAUGCUGUAGGUGCCCCAAAC____AGCUGAGGUACUGAGUAUAGCAUCUAGAAAUAUUUAAAAA_CUUUGUAGCUGUCGCAAAUACGGCACAAUU__G_A_________ .........((((((((((((((((.((..........)).))))))...))))))))))............................................................ (-16.74 = -17.26 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:20 2011