
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,568,501 – 16,568,605 |
| Length | 104 |
| Max. P | 0.999548 |

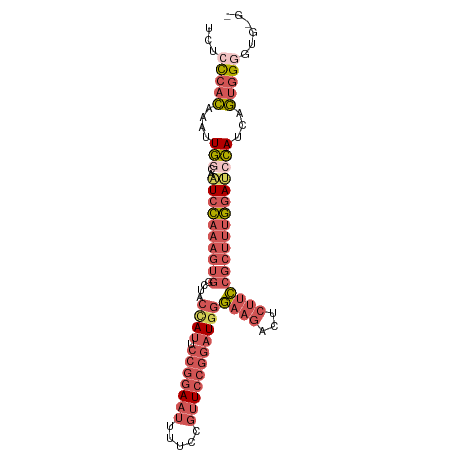
| Location | 16,568,501 – 16,568,595 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.56305 |
| G+C content | 0.48791 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -20.26 |
| Energy contribution | -23.66 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
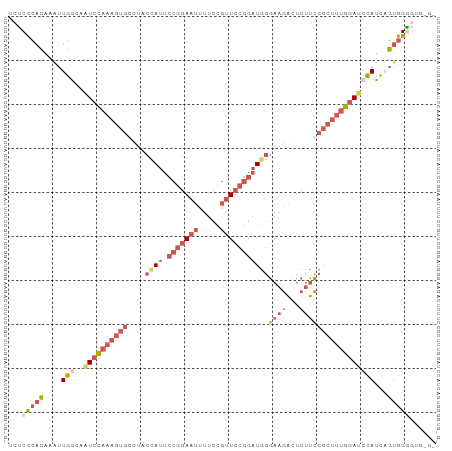
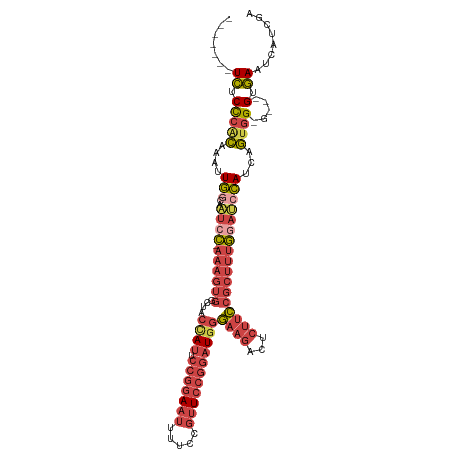
>dm3.chrX 16568501 94 + 22422827 UCUCCCACAAAUUGACAAUCCAAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGGGGU--- .(((((((....(((..(((((((((((...((((.(((((((.....)))))))))))((....))..)))))))))))...)))))))))).--- ( -41.40, z-score = -4.24, R) >droAna3.scaffold_13334 970066 91 + 1562580 UUUCUAAAAACCUAGUACUUUCCA------ACCAGGGCUGAAAGUCCUCCUACUGGUUCUAACUUCCAAGUGACUCAUAGCUAUUUUUGGGCCUGGG ........................------.(((((.(..(((((....(((..((((...(((....)))))))..)))..)))))..).))))). ( -19.90, z-score = 0.09, R) >droEre2.scaffold_4690 6926839 92 + 18748788 UCCCCCACAAGUUGACAAUCCAAAGUGAGUUCUAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGAUUCUUCCGCUUUGGAGCCAUCAGCGGGU----- ...(((....(((((...((((((((.........((((((((.....))))))))..(((((...)))))))))))))....)))))))).----- ( -34.20, z-score = -3.03, R) >droSec1.super_17 391158 97 + 1527944 UCUCCCACAAAUUGGCAAUCCAAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCUGUGGAGUGUGU ....(((((...(((..(((((((((((...((((.(((((((.....)))))))))))((....))..))))))))))))))..)))))....... ( -39.70, z-score = -4.13, R) >droSim1.chrX 12736629 97 + 17042790 UCUCCCACAAAUUGGCAAUCCAAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGAGUGGGU ...(((((..(((((..(((((((((((...((((.(((((((.....)))))))))))((....))..)))))))))))...)))))...))))). ( -41.10, z-score = -3.74, R) >consensus UCUCCCACAAAUUGGCAAUCCAAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGGGUG_G_ ...(((((....(((..((((((((((....((((.(((((((.....)))))))))))((((...)))))))))))))))))...)))))...... (-20.26 = -23.66 + 3.40)

| Location | 16,568,501 – 16,568,595 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.56305 |
| G+C content | 0.48791 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.98 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.989996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16568501 94 - 22422827 ---ACCCCCACUGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCACUUUGGAUUGUCAAUUUGUGGGAGA ---...((((((((..(.(((((((.((..........((((((((((.......)))))).))))...)).))))))))..)))....)))))... ( -35.12, z-score = -2.25, R) >droAna3.scaffold_13334 970066 91 - 1562580 CCCAGGCCCAAAAAUAGCUAUGAGUCACUUGGAAGUUAGAACCAGUAGGAGGACUUUCAGCCCUGGU------UGGAAAGUACUAGGUUUUUAGAAA ................(((..(((((.(((((.........)))..))...)))))..)))((((((------........)))))).......... ( -17.30, z-score = 1.61, R) >droEre2.scaffold_4690 6926839 92 - 18748788 -----ACCCGCUGAUGGCUCCAAAGCGGAAGAAUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUAGAACUCACUUUGGAUUGUCAACUUGUGGGGGA -----.((((((((..(.(((((((.(((((...)))))..(((((((.......)))))))..........))))))))..)))....)))))... ( -34.40, z-score = -2.61, R) >droSec1.super_17 391158 97 - 1527944 ACACACUCCACAGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCACUUUGGAUUGCCAAUUUGUGGGAGA .....((((((((((((((((((((.((..........((((((((((.......)))))).))))...)).))))))))..)).)))))).)))). ( -36.72, z-score = -3.08, R) >droSim1.chrX 12736629 97 - 17042790 ACCCACUCCACUGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCACUUUGGAUUGCCAAUUUGUGGGAGA .(((((((((....))))(((((((.((..........((((((((((.......)))))).))))...)).)))))))..........)))))... ( -34.32, z-score = -1.92, R) >consensus _C_AACCCCACUGAUGGAUCCAAAGCGGAAGAGUCUUCCCAUCCGGAACGGAAAAUUCCGGAAUGGUAGCCACUUUGGAUUGCCAAUUUGUGGGAGA ......(((((...(((((((((((.(((((...)))))..(((((((.......)))))))..........))))))))..)))....)))))... (-20.74 = -21.98 + 1.24)

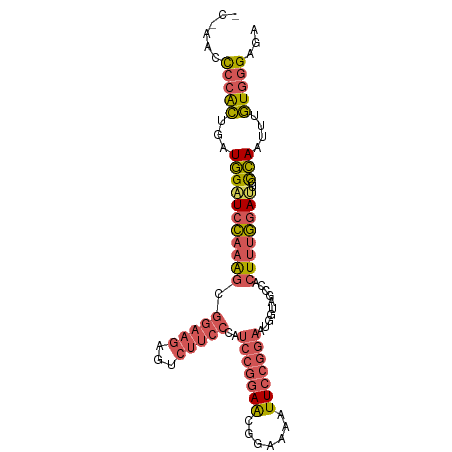
| Location | 16,568,501 – 16,568,605 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.73 |
| Shannon entropy | 0.51886 |
| G+C content | 0.47823 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -16.88 |
| Energy contribution | -19.47 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
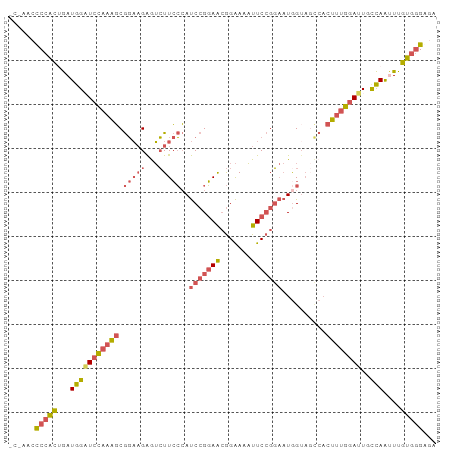
>dm3.chrX 16568501 104 + 22422827 -------UCUCCCACAAAUUGACAAUCC-AAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGGG---GUGAAUCAUCGA -------.(((((((....(((..((((-(((((((...((((.(((((((.....)))))))))))((....))..)))))))))))...)))))))))---)........... ( -41.40, z-score = -3.28, R) >droAna3.scaffold_13334 970066 101 + 1562580 -------UUUCUAAAAACCUAGUACUUU-------CCAACCAGGGCUGAAAGUCCUCCUACUGGUUCUAACUUCCAAGUGACUCAUAGCUAUUUUUGGGCCUGGGAACUUAUCAA -------..........(((((.....(-------((((....(((((..((((((.....(((.........))))).))))..)))))....))))).))))).......... ( -20.70, z-score = 0.27, R) >droEre2.scaffold_4690 6926839 102 + 18748788 -------UCCCCCACAAGUUGACAAUCC-AAAGUGAGUUCUAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGAUUCUUCCGCUUUGGAGCCAUCAGCGG-----GUGAAUCAUCGG -------.....(((..(((((...(((-(((((.........((((((((.....))))))))..(((((...)))))))))))))....)))))..-----)))......... ( -36.70, z-score = -2.66, R) >droYak2.chrX 10675522 110 + 21770863 UUUGUUCUCUCCCGCAAAUUGGCAAUCCCAAAGUGGCUUCUAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGAUUCUUCCGCUUUGCCUCCAUCAGUGG-----GUGAAUCAUCGA ...((((...(((((....(((......(((((((((((((..((((((((.....))))))))..)))))......))))))))...)))...))))-----).))))...... ( -37.00, z-score = -2.59, R) >droSec1.super_17 391158 107 + 1527944 -------UCUCCCACAAAUUGGCAAUCC-AAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCUGUGGAGUGUGUGAAUCAUCGA -------....(((((...(((..((((-(((((((...((((.(((((((.....)))))))))))((....))..))))))))))))))..)))))................. ( -39.70, z-score = -3.08, R) >droSim1.chrX 12736629 107 + 17042790 -------UCUCCCACAAAUUGGCAAUCC-AAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGGAGUGGGUGAAUCAUCGA -------((.(((((..(((((..((((-(((((((...((((.(((((((.....)))))))))))((....))..)))))))))))...)))))...))))).))........ ( -43.40, z-score = -3.42, R) >consensus _______UCUCCCACAAAUUGGCAAUCC_AAAGUGGCUACCAUUCCGGAAUUUUCCGUUCCGGAUGGGAAGACUCUUCCGCUUUGGAUCCAUCAGUGG_G___GUGAAUCAUCGA ...........((((....(((..((((.((((((....((((.(((((((.....)))))))))))((((...)))))))))))))))))...))))................. (-16.88 = -19.47 + 2.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:16 2011