| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,568,358 – 16,568,472 |
| Length | 114 |
| Max. P | 0.536136 |

| Location | 16,568,358 – 16,568,472 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Shannon entropy | 0.45009 |
| G+C content | 0.51029 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.536136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
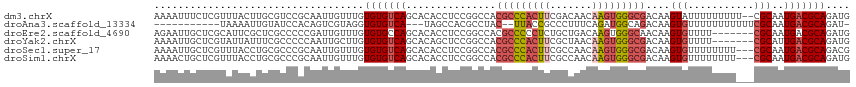
>dm3.chrX 16568358 114 + 22422827 AAAAUUUCUCGUUUACUUGCGUCCGCAAUUGUUUGUGUGUCAGCACACCUCCGGCCACGCCCACUUCGACAACAAGUGGGCGACAAGUAUUUUUUUUU--CGCAAUGACGCAGAUG ................(((((((.((........(((((....))))).........(((((((((.......)))))))))................--.))...)))))))... ( -33.10, z-score = -2.19, R) >droAna3.scaffold_13334 969942 99 + 1562580 -----------UAAAAUUGUAUCCACAGUCGUAGGUGUGUCA---UAGCCACGCCUAC--UUACCGCCCUUUCAGAUGGCAGACAAGUGUUUUUUUUUUUCGCAAUGACGCAGAU- -----------.....(((..((....((((((((((((...---....)))))))))--.....(((.........))).)))..(((...........)))...))..)))..- ( -24.50, z-score = -1.91, R) >droEre2.scaffold_4690 6926703 109 + 18748788 AGAAUUGCUCGCAUUCGCUCGCCCCCGAUUGUUUGUGUGCCAGCACACCUCCGGCCACGCCCCCUCUGCUGACAAGUGGGCAACAAGUGUUUU-------CGCAAUGACGCAGAUG ....((((((((..(((........)))(((((.(((((....)))))....(((...))).........)))))))))))))...(((((..-------......)))))..... ( -26.80, z-score = 1.12, R) >droYak2.chrX 10675372 109 + 21770863 AAAAUUGCUCGUAUUAUUUCGCCCCCAAUUGCUUGUGUGUCAGCACAGCUCCGGCCACGCCCACUUCGCUAACAAGUGGGCGACAAGUGUUUU-------CGCAUUGACGCAGAUG ....((((((((........(((.......(((.((((....)))))))...)))..(((((((((.......)))))))))))..(((....-------)))...)).))))... ( -31.70, z-score = -1.26, R) >droSec1.super_17 391016 113 + 1527944 AAAAUUGCUCGUUUACCUGCGCCCGCAAUUGUUUGUGUGUCAGCACACCUCCGGCCACGCCCACUUCGCCAACAAGUGGGCGACAAGUGUUUUUUUU---CGCAAUGACGCAGACG ..((((((.(((......)))...))))))(((((((((((.((.(......)))...((((((((.......)))))))))))..(((........---)))....)))))))). ( -35.40, z-score = -1.62, R) >droSim1.chrX 12736484 113 + 17042790 AAAACUGCUCGUUUACCUGCGCCCGCAAUUGUUUGUGUGUCAGCACACCUCCGGCCACGCCCACUUCGCCAACAAGUGGGCGACAAGUGUUUUUUUU---CGCAAUGACGCAGAUG ....(((((((((....(((((((((..(((((.(((((....)))))....(((............))))))))))))))).)).(((........---)))))))).))))... ( -36.00, z-score = -1.76, R) >consensus AAAAUUGCUCGUAUACUUGCGCCCGCAAUUGUUUGUGUGUCAGCACACCUCCGGCCACGCCCACUUCGCCAACAAGUGGGCGACAAGUGUUUUUUUU___CGCAAUGACGCAGAUG ...................................(((((((...............(((((((((.......)))))))))....(((...........)))..))))))).... (-17.84 = -18.78 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:13 2011