
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,567,261 – 16,567,372 |
| Length | 111 |
| Max. P | 0.984952 |

| Location | 16,567,261 – 16,567,372 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.18 |
| Shannon entropy | 0.49590 |
| G+C content | 0.36673 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16567261 111 - 22422827 AUCCGGUAAUCGUACUUAAUAAAUGUGGAAACACCAAAUUGCAUAAAACUAAUGCAAUUUGUUGAGUCUUUCUGUUGCAUAAGCCAGAGCUAGAAUACUCUUACUUCAUUU--------- ....(((((..((((((((.....(((....)))((((((((((.......)))))))))))))))((((((((..((....)))))))..))).)))..)))))......--------- ( -25.80, z-score = -2.82, R) >droEre2.scaffold_4690 6925695 110 - 18748788 -UCAAGAAAAAGCACU---UCAAUGUGGAAACACCAAAUUGCAUAAAAGUACUGCAAUUCGUUGUUUUCUGUU---GCAUAACCC--AGCUGGAAGACUU-CAAAUGGCUUCAAUGGCUC -...(((((((((...---.....(((....)))..(((((((.........))))))).))).))))))...---.........--((((.((((.((.-.....))))))...)))). ( -22.20, z-score = -0.06, R) >droYak2.chrX 10674370 107 - 21770863 AUCAAGCUAAAGUACU---UGAAUGUGGAAACUCCAAAUUGCUUAAAAGUUGUGCAAUUUGUUGAGUUUUCCA---CCA-----C--AGCAAGAAUACUUGUAAAUAGCUUCAGAUUCUC ...(((((((((((((---((...((((((((((((((((((...........))))))))..))).))))))---)..-----.--..))))..))))).....))))))......... ( -29.50, z-score = -3.46, R) >droSec1.super_17 389975 110 - 1527944 AUCCGGUAAUCGUACU---UAAAUGUGGAAACACCGAAUUGCGUAAAACUAAUGCAAUUUGUUGAGUUUUUCUGUUGCAUAAGCC--AGCUAGAAUGCUCUUACUUCAUCCACAU----- ....((((....))))---...(((((((.....((((((((((.......))))))))))..((((..(((((((((....).)--))).)))).))))........)))))))----- ( -28.50, z-score = -3.10, R) >droSim1.chrX 12735451 110 - 17042790 AUCCGGUAAUCGUACU---UAAAUGUGGAAACACCGAAUUGCGUAAAACUAAUGCAAUUUGUUGAGUUUUUCUGUUGCAUAAGCC--AGCUAGAAUGCUCUUACUCCACCCAUCU----- ....(((.........---.....(((....)))((((((((((.......))))))))))..((((..(((((((((....).)--))).)))).)))).......))).....----- ( -26.30, z-score = -2.61, R) >consensus AUCCGGUAAUCGUACU___UAAAUGUGGAAACACCAAAUUGCAUAAAACUAAUGCAAUUUGUUGAGUUUUUCUGUUGCAUAAGCC__AGCUAGAAUACUCUUACUUCACUUAAAU_____ .((.(((.................(((....)))((((((((((.......))))))))))...........................))).)).......................... (-13.36 = -13.88 + 0.52)

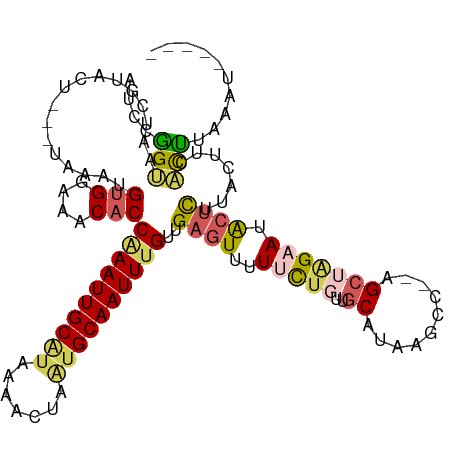
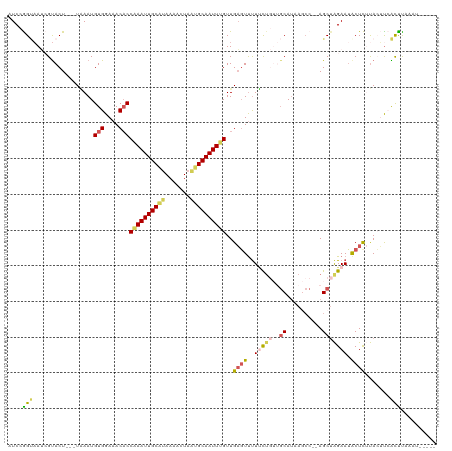
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:12 2011