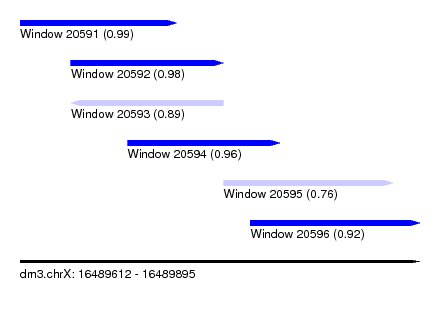
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,489,612 – 16,489,895 |
| Length | 283 |
| Max. P | 0.985446 |

| Location | 16,489,612 – 16,489,723 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Shannon entropy | 0.54099 |
| G+C content | 0.46374 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -13.92 |
| Energy contribution | -15.95 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
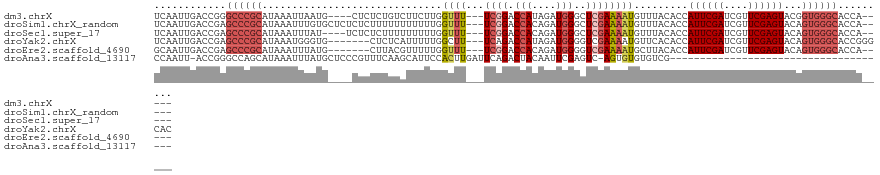
>dm3.chrX 16489612 111 + 22422827 UCAAUUGACCGGGCCCGCAUAAAUUAAUG----CUCUCUGUCUUCUUGGUUU---UCGGACCAUAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACGGUGGGCACCA----- .........(((((((((((......)))----).....((((...(((((.---...))))).)))))))))))....(((((((...((((((....))))))...)))))))...----- ( -37.40, z-score = -2.68, R) >droSim1.chrX_random 4327174 115 + 5698898 UCAAUUGACCGAGCCCGCAUAAAUUUGUGCUCUCUCUUUUUUUUUUUGGUUU---UCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACCA----- .........((((((((((((....)))))............(((.(((((.---...))))).))).)))))))....(((((((...((((((....))))))...)))))))...----- ( -35.00, z-score = -2.78, R) >droSec1.super_17 312951 111 + 1527944 UCAAUUGACCGAGCCCGCAUAAAUUUAU----UCUCUCUUUUUUUUUGGUUU---UCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACCA----- .........((((((((.(((....)))----.)........(((.(((((.---...))))).))).)))))))....(((((((...((((((....))))))...)))))))...----- ( -28.40, z-score = -1.58, R) >droYak2.chrX 10595696 113 + 21770863 UCAAUUGACCGAGCCCGCAUAAAUGGGUG-------CUCUCAUUUUUGGCUU---UCAGACCAUAGAUGGGGUCGAAAAUGUUCACACCAUUCGAUCGUUCGAGUACAGUGGGCACCGGGCAC .....((.(((.((((((.......((((-------....(((((((((((.---.....(((....))))))))))))))....))))((((((....))))))...))))))..))).)). ( -39.40, z-score = -2.52, R) >droEre2.scaffold_4690 6848245 108 + 18748788 GCAAUUGACCGAGCCCGCAUAAAUUUAUG-------CUUACGUUUUUGGUUU---UCGGACCACAGAUGGGGUCGAAAAUGCUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACCA----- .........(((.((((((((....))))-------)...(((((.(((((.---...))))).)))))))))))....(((((((...((((((....))))))...)))))))...----- ( -34.70, z-score = -2.64, R) >droAna3.scaffold_13117 460172 84 + 5790199 CCAAUU-ACCGGGCCAGCAUAAAUUUAUGCUCCCGUUUCAAGCAUUCCACUUGAUUCAGACUACAAUUCGAGUC-AGUGUGUGUCG------------------------------------- ......-..((((..((((((....))))))))))......((((..((((.(((((.((.......)))))))-)))).))))..------------------------------------- ( -21.50, z-score = -2.94, R) >consensus UCAAUUGACCGAGCCCGCAUAAAUUUAUG____CUCUUUUUGUUUUUGGUUU___UCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACCA_____ .........(((.(((((((......))).................((((((.....))))))....)))).)))....(((((((...((((((....))))))...)))))))........ (-13.92 = -15.95 + 2.03)
| Location | 16,489,648 – 16,489,756 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Shannon entropy | 0.35798 |
| G+C content | 0.48447 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -21.36 |
| Energy contribution | -24.24 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
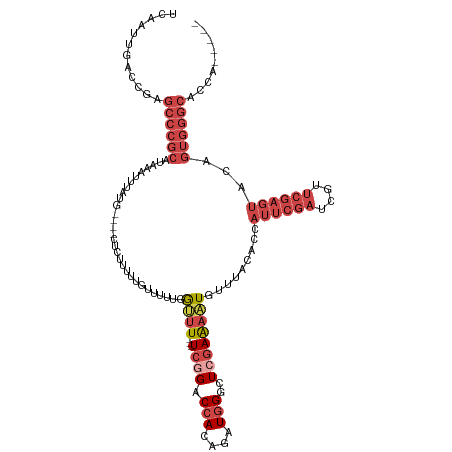
>dm3.chrX 16489648 108 + 22422827 UCUUCUUGGUUUUCGGACCAUAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACGGUGGGCACC-------ACAGACUUUUACGGCCCACUGUACGUUCCACCAG ......((((..((((((.((.(((((.................)))))..)).))))))(((((((((((...-------.............))))))))))).....)))). ( -33.52, z-score = -2.16, R) >droSim1.chrX_random 4327214 108 + 5698898 UUUUUUUGGUUUUCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACC-------AAAGACUUUUACGGCCCACUGUACGUUUCACCAG ......(((((((((..((.(....)))..))))))((...((.....((((....))))(((((((((((...-------(((....)))...)))))))))))))..))))). ( -34.80, z-score = -3.49, R) >droSec1.super_17 312987 108 + 1527944 UUUUUUUGGUUUUCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACC-------AAAGACUUUUAGGGCCCACUGUACGUUUCACCAG ......(((((((((..((.(....)))..))))))((...((.....((((....))))(((((((((((.((-------...........)))))))))))))))..))))). ( -38.10, z-score = -4.24, R) >droYak2.chrX 10595731 113 + 21770863 --UUUUUGGCUUUCAGACCAUAGAUGGGGUCGAAAAUGUUCACACCAUUCGAUCGUUCGAGUACAGUGGGCACCGGGCACCAAUGGCUUUAAUGGCCCCCUGUUGGUUGCACCAG --.....((.((((.((((........)))))))).(((((((...((((((....))))))...)))))))))((((((((((((((.....))))....))))).))).)).. ( -37.70, z-score = -1.33, R) >droEre2.scaffold_4690 6848280 89 + 18748788 --UUUUUGGUUUUCGGACCACAGAUGGGGUCGAAAAUGCUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACC----ACCGUAGGUUUAACCAG-------------------- --....(((((...(((((....(((((((.......((((((...((((((....))))))...)))))))))----.)))).)))))))))).-------------------- ( -26.41, z-score = -1.74, R) >consensus U_UUUUUGGUUUUCGGACCACAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCACC_______AAAGACUUUUACGGCCCACUGUACGUUUCACCAG ........((((((((.(((....)))..))))))))...........((((....))))(((((((((((.......................))))))))))).......... (-21.36 = -24.24 + 2.88)

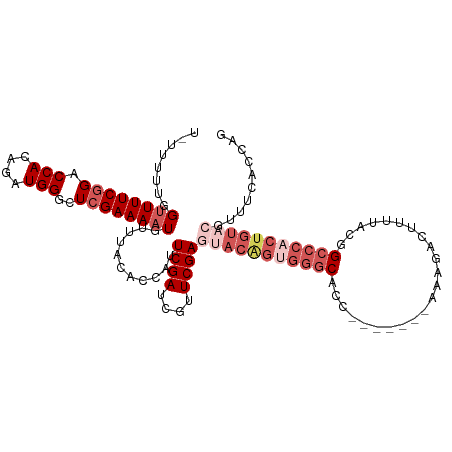
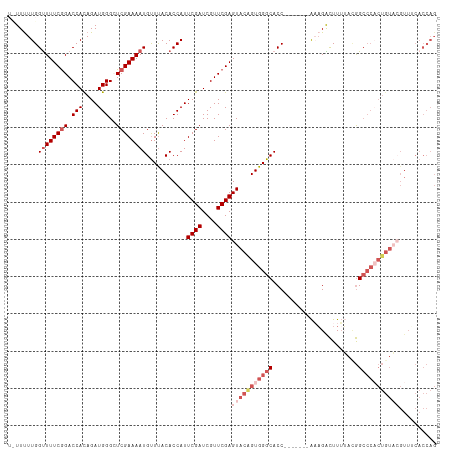
| Location | 16,489,648 – 16,489,756 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Shannon entropy | 0.35798 |
| G+C content | 0.48447 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -19.89 |
| Energy contribution | -22.21 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16489648 108 - 22422827 CUGGUGGAACGUACAGUGGGCCGUAAAAGUCUGU-------GGUGCCCACCGUACUCGAACGAUCGAAUGGUGUAAACAUUUUCGAGCCCAUCUAUGGUCCGAAAACCAAGAAGA .(((((((.((((..(((((((((...((((.((-------((...)))).).)))...))).(((((.((((....))))))))))))))).)))).)))....))))...... ( -31.60, z-score = -1.01, R) >droSim1.chrX_random 4327214 108 - 5698898 CUGGUGAAACGUACAGUGGGCCGUAAAAGUCUUU-------GGUGCCCACUGUACUCGAACGAUCGAAUGGUGUAAACAUUUUCGAGCCCAUCUGUGGUCCGAAAACCAAAAAAA .((((...(((((((((((((..((((....)))-------)..)))))))))))((((....))))...))........(((((..(((....).))..)))))))))...... ( -35.50, z-score = -3.24, R) >droSec1.super_17 312987 108 - 1527944 CUGGUGAAACGUACAGUGGGCCCUAAAAGUCUUU-------GGUGCCCACUGUACUCGAACGAUCGAAUGGUGUAAACAUUUUCGAGCCCAUCUGUGGUCCGAAAACCAAAAAAA .((((...(((((((((((((.(((((....)))-------)).)))))))))))((((....))))...))........(((((..(((....).))..)))))))))...... ( -39.10, z-score = -4.62, R) >droYak2.chrX 10595731 113 - 21770863 CUGGUGCAACCAACAGGGGGCCAUUAAAGCCAUUGGUGCCCGGUGCCCACUGUACUCGAACGAUCGAAUGGUGUGAACAUUUUCGACCCCAUCUAUGGUCUGAAAGCCAAAAA-- .(((((((.(((((((.(((((......(((...))).....).)))).))))..((((....)))).))))))......((((((((........)))).))))))))....-- ( -32.20, z-score = -0.04, R) >droEre2.scaffold_4690 6848280 89 - 18748788 --------------------CUGGUUAAACCUACGGU----GGUGCCCACUGUACUCGAACGAUCGAAUGGUGUAAGCAUUUUCGACCCCAUCUGUGGUCCGAAAACCAAAAA-- --------------------.((.(((.(((((((((----((...)))))))).((((....))))..))).))).))(((((((((.(....).))).)))))).......-- ( -24.80, z-score = -1.64, R) >consensus CUGGUGAAACGUACAGUGGGCCGUAAAAGUCUUU_______GGUGCCCACUGUACUCGAACGAUCGAAUGGUGUAAACAUUUUCGAGCCCAUCUGUGGUCCGAAAACCAAAAA_A ..........((((((((((((....................).)))))))))))((((....)))).(((((....)..(((((..((.......))..)))))))))...... (-19.89 = -22.21 + 2.32)

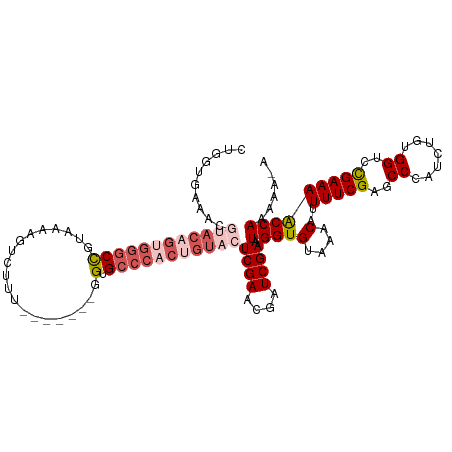
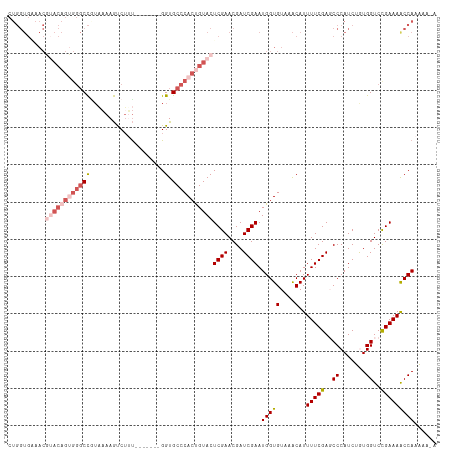
| Location | 16,489,688 – 16,489,796 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.28768 |
| G+C content | 0.49019 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -22.44 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16489688 108 + 22422827 UACACCAUUCGAUCGUUCGAGUACGGUGGGCACC-------ACAGACUUUUACGGCCCACUGUACGUUCCACCAGUUAGUCAGUCAGUCAUCAAAACAUUGCCAAUUAGUGGGAG ........((((....))))(((((((((((...-------.............))))))))))).((((((.((((.(.((((.............))))).)))).)))))). ( -31.61, z-score = -2.73, R) >droSim1.chrX_random 4327254 101 + 5698898 UACACCAUUCGAUCGUUCGAGUACAGUGGGCACC-------AAAGACUUUUACGGCCCACUGUACGUUUCACCAGUCAGUCG-------AUCAAAACAUUGCCAAUUAGUGGGAG ........((((....))))(((((((((((...-------(((....)))...))))))))))).(..(((.(((..(.((-------((......)))))..))).)))..). ( -29.50, z-score = -2.86, R) >droSec1.super_17 313027 101 + 1527944 UACACCAUUCGAUCGUUCGAGUACAGUGGGCACC-------AAAGACUUUUAGGGCCCACUGUACGUUUCACCAGUCAGUCG-------AUUAAAACAUUGCCAAUUAGUGGGAG ........((((....))))(((((((((((.((-------...........))))))))))))).(..(((.(((..(.((-------((......)))))..))).)))..). ( -32.80, z-score = -3.82, R) >droYak2.chrX 10595769 102 + 21770863 CACACCAUUCGAUCGUUCGAGUACAGUGGGCACCGGGCACCAAUGGCUUUAAUGGCCCCCUGUUGGUUGCACCAG-----CG------CAUCAAAAUAUUGCCAAUUUCAGGG-- ....((((((((....))))))....(((((.(.((((((((((((((.....))))....))))).))).)).)-----.)------)..(((....))))))......)).-- ( -28.00, z-score = 0.22, R) >consensus UACACCAUUCGAUCGUUCGAGUACAGUGGGCACC_______AAAGACUUUUACGGCCCACUGUACGUUUCACCAGUCAGUCG_______AUCAAAACAUUGCCAAUUAGUGGGAG ....((((((((....))))(((((((((((.......................)))))))))))...........................................))))... (-22.44 = -23.50 + 1.06)

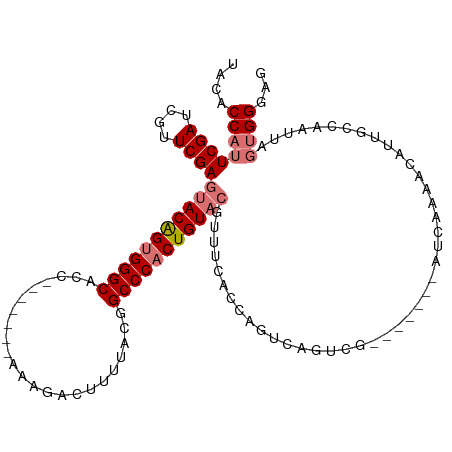
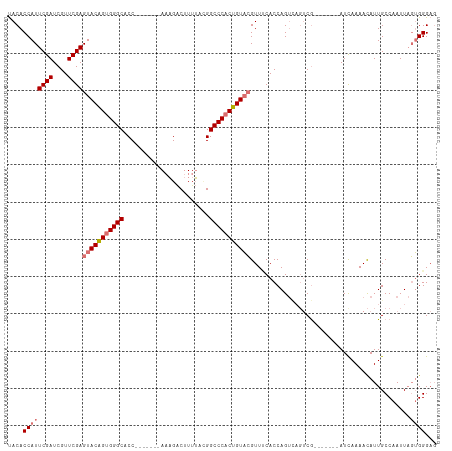
| Location | 16,489,756 – 16,489,876 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 75.05 |
| Shannon entropy | 0.39740 |
| G+C content | 0.47742 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -15.40 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.755451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
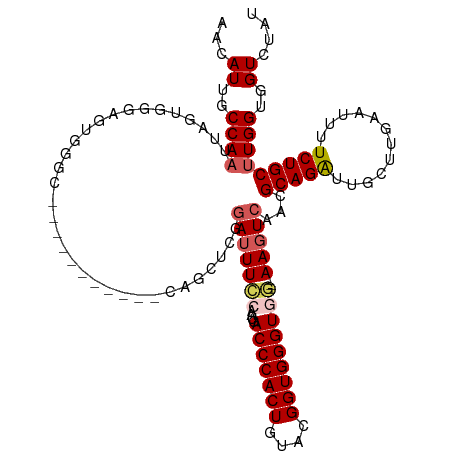
>dm3.chrX 16489756 120 + 22422827 UUAGUCAGUCAGUC-AUCAAAACAUUGCCAAUUAGUGGGAGUGGGCAAGGGCUGCGCCAGUUCGGAUUUCCAUUGACCCACUGUACGGUGGGUGGGGGAAGUCAAUGCAGAUUGCUUGAAU ....((((.(((((-.......((((.(((.....))).)))).(((.((((((...)))))).(((((((.((.(((((((....))))))).)))))))))..))).))))).)))).. ( -46.10, z-score = -3.11, R) >droSim1.chrX_random 4327322 99 + 5698898 --------UCAGUCGAUCAAAACAUUGCCAAUUAGUGGGAGUGGGC-----------CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG---AAGUCAACGCAGAUUGCUUGAAU --------((((.(((((....((((.(((.....))).))))...-----------..((...(((((((....(((((((....)))))))))---)))))...)).))))).)))).. ( -36.90, z-score = -2.84, R) >droSec1.super_17 313095 99 + 1527944 --------UCAGUCGAUUAAAACAUUGCCAAUUAGUGGGAGUGGGC-----------CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG---AAGUCAACGCAGAUUGCUUGAAU --------((((.(((((....((((.(((.....))).))))...-----------..((...(((((((....(((((((....)))))))))---)))))...)).))))).)))).. ( -34.40, z-score = -2.34, R) >droYak2.chrX 10595844 94 + 21770863 ------------CGCAUCAAAAUAUUGCCAAUUU-CAGGGCUCCGC-----------CAGCUCGAAUUUCAAAUGACCCACUGUACGGUGGGUGA---AAGUCAGCGCAGAUUGCUUCAAU ------------.(((((........(((.....-...)))...((-----------..(((.((.((((.....(((((((....)))))))))---)).))))))).)).)))...... ( -25.30, z-score = -0.92, R) >droEre2.scaffold_4690 6848369 91 + 18748788 ------------UCCAUCAAAACAUUGCCAAUUUCAAGGCAAGAGC---------------UCGGAUUUUAAAUAACCCACUGUAAGGUGGGUGA---AACUCUGCGCAGGUUGCUUGAAU ------------....(((((((.(((((........)))))..((---------------.((((.(((.....(((((((....))))))).)---)).)))).))..)))..)))).. ( -27.10, z-score = -2.23, R) >consensus ________UCAGUCCAUCAAAACAUUGCCAAUUAGUGGGAGUGGGC___________CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG___AAGUCAACGCAGAUUGCUUGAAU ........................((((..............((((.............)))).(((((....(.(((((((....))))))).)...)))))...))))........... (-15.40 = -16.12 + 0.72)
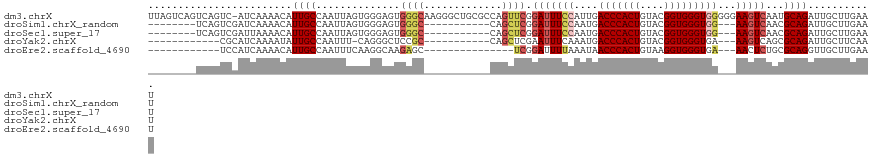
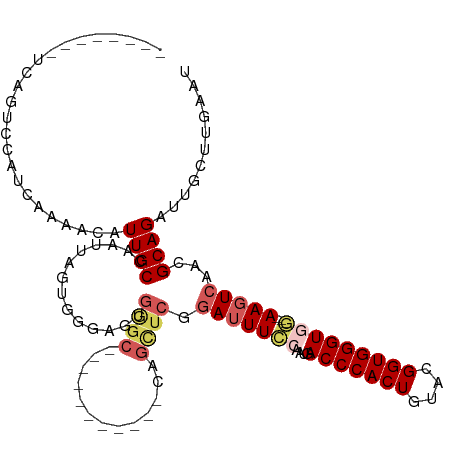
| Location | 16,489,775 – 16,489,895 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Shannon entropy | 0.31103 |
| G+C content | 0.48494 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16489775 120 + 22422827 AACAUUGCCAAUUAGUGGGAGUGGGCAAGGGCUGCGCCAGUUCGGAUUUCCAUUGACCCACUGUACGGUGGGUGGGGGAAGUCAAUGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUAC ...((..((((......(((.((((((.....))).))).))).(((((((.((.(((((((....))))))).)))))))))...(((((............)))))))))..)).... ( -45.20, z-score = -2.14, R) >droSim1.chrX_random 4327334 106 + 5698898 AACAUUGCCAAUUAGUGGGAGUGGGC-----------CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG---AAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUUU ..((((.(((.....))).))))(((-----------((.(...(((((((....(((((((....)))))))))---)))))...(((((............)))))..).)))))... ( -40.50, z-score = -2.73, R) >droSec1.super_17 313107 106 + 1527944 AACAUUGCCAAUUAGUGGGAGUGGGC-----------CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG---AAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUAU ..((((.(((.....))).))))(((-----------((.(...(((((((....(((((((....)))))))))---)))))...(((((............)))))..).)))))... ( -40.50, z-score = -2.78, R) >droYak2.chrX 10595852 105 + 21770863 AAUAUUGCCAAUUUCAGGGCUCCGCC------------AGCUCGAAUUUCAAAUGACCCACUGUACGGUGGGUGA---AAGUCAGCGCAGAUUGCUUCAAUUGUCUGCCUGGUGGUCUAU ......(((........))).(((((------------((((.((.((((.....(((((((....)))))))))---)).)))))((((((..........)))))).))))))..... ( -36.50, z-score = -2.52, R) >droEre2.scaffold_4690 6848377 102 + 18748788 AACAUUGCCAAUUUCAAGG---CAAG------------AGCUCGGAUUUUAAAUAACCCACUGUAAGGUGGGUGA---AACUCUGCGCAGGUUGCUUGAAUUGUCUGCUUGGUGGUCUAU ...((..((((.((((((.---(((.------------.((.((((.(((.....(((((((....))))))).)---)).)))).))...)))))))))........))))..)).... ( -30.00, z-score = -1.32, R) >consensus AACAUUGCCAAUUAGUGGGAGUGGGC___________CAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUGG___AAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUAU ...((..((((.................................(((((....(.(((((((....))))))).)...)))))...(((((............)))))))))..)).... (-25.20 = -25.40 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:06 2011