| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,486,968 – 16,487,029 |
| Length | 61 |
| Max. P | 0.598260 |

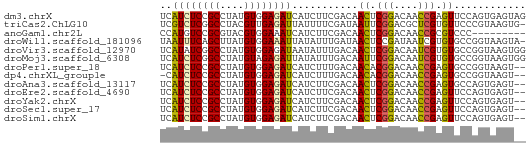
| Location | 16,486,968 – 16,487,029 |
|---|---|
| Length | 61 |
| Sequences | 13 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 82.80 |
| Shannon entropy | 0.38907 |
| G+C content | 0.49142 |
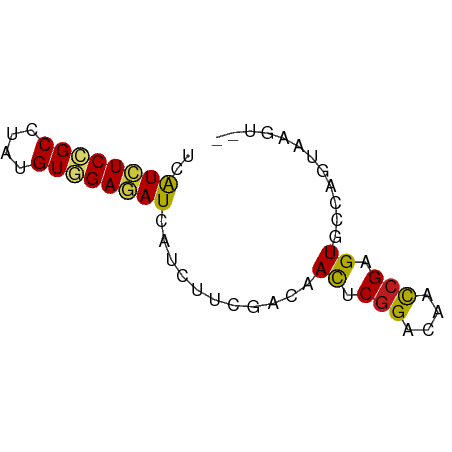
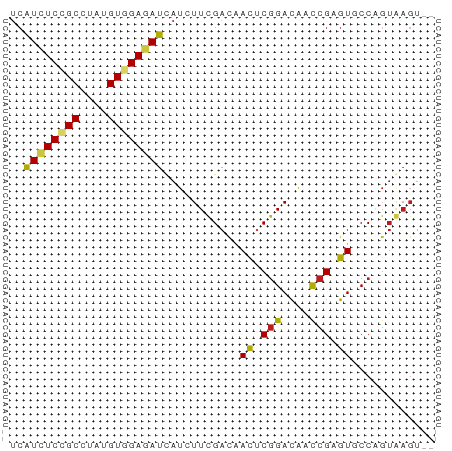
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.25 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16486968 61 - 22422827 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUUCCAGUGAGUAG ..((((((((....))))))))....((((..(((((((....)))))))....))))... ( -20.40, z-score = -2.64, R) >triCas2.ChLG10 8277312 60 - 8806720 UCGUCUCGGCCUACGUUGAGAUUAUUUUCGAUAAUUCGGACGCUCGUGUUCCCGUAAGUG- ..((((((((....))))))))....(((((....)))))((((..((....))..))))- ( -14.60, z-score = -0.75, R) >anoGam1.chr2L 18320010 52 + 48795086 CCAUGUCCGCGUACGUGGAAAUCAUCUUCGACAACUCGGACAACCGCGUCCC--------- ...((((((.((..((.(((......))).)).)).))))))..........--------- ( -13.80, z-score = -1.28, R) >droWil1.scaffold_181096 1245506 60 - 12416693 UAAUUUCAGCUUAUGUGGAAAUUAUAUUUGAUAACUCCGAUAAUCGUGUGCCGGUAAGUA- (((((((.((....)).))))))).........((((((............)))..))).- ( -7.40, z-score = 0.51, R) >droVir3.scaffold_12970 10755881 61 + 11907090 UCAUAUCGGCCUAUGUGGAGAUAAUAUUUGACAACUCGGACAAUCGUGUGCCGGUAAGUGG ...(((((((...(((.(((......))).)))((.((......)).)))))))))..... ( -12.90, z-score = 0.06, R) >droMoj3.scaffold_6308 1245722 61 - 3356042 UCAUCUCGGCCUAUGUAGAGAUUAUAUUUGACAAUUCGGACAAUCGUGUGCCGGUAAGUGG ..(((((.((....)).))))).........((..(((((((....))).))))....)). ( -10.60, z-score = 0.69, R) >droPer1.super_18 1856874 59 - 1952607 UCAUCUCCGCCUAUGUGGAGAUCAUCUUUGACAACACGGACAACCGAGUGCCGGUAAGU-- ..((((((((....))))))))..(((.((....)).)))..((((.....))))....-- ( -15.30, z-score = -0.88, R) >dp4.chrXL_group1e 2958304 58 - 12523060 -CAUCUCCGCCUAUGUGGAGAUCAUCUUUGACAACACGGACAACCGAGUGCCGGUAAGU-- -.((((((((....))))))))..(((.((....)).)))..((((.....))))....-- ( -15.30, z-score = -0.97, R) >droAna3.scaffold_13117 439887 59 + 5790199 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUGCCAGUGAGU-- ..((((((((....))))))))....((((...((((((....)))))).....)))).-- ( -20.30, z-score = -2.36, R) >droEre2.scaffold_4690 6845235 59 - 18748788 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUUCCAGUGAGU-- ..((((((((....))))))))....((((..(((((((....)))))))....)))).-- ( -20.40, z-score = -2.62, R) >droYak2.chrX 10593001 59 - 21770863 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUUCCAGUGAGU-- ..((((((((....))))))))....((((..(((((((....)))))))....)))).-- ( -20.40, z-score = -2.62, R) >droSec1.super_17 310231 59 - 1527944 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUUCCAGUGAGU-- ..((((((((....))))))))....((((..(((((((....)))))))....)))).-- ( -20.40, z-score = -2.62, R) >droSim1.chrX 12682112 59 - 17042790 UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUUCCAGUGAGU-- ..((((((((....))))))))....((((..(((((((....)))))))....)))).-- ( -20.40, z-score = -2.62, R) >consensus UCAUCUCCGCCUAUGUGGAGAUCAUCUUCGACAACUCGGACAACCGAGUGCCAGUAAGU__ ..((((((((....))))))))...........((.(((....))).))............ (-10.42 = -10.25 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:50:00 2011