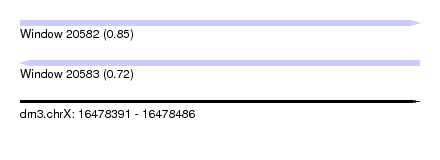
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,478,391 – 16,478,486 |
| Length | 95 |
| Max. P | 0.853857 |

| Location | 16,478,391 – 16,478,486 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.35835 |
| G+C content | 0.40852 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
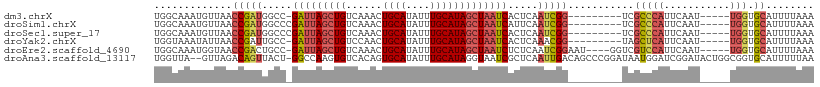
>dm3.chrX 16478391 95 + 22422827 UGGCAAAUGUUAACCGAUGGCC-GAUUAGCUGUCAAACUGCAUAUUUGCAUAGCUAAUCACUCAAUCGG---------UCGCCCAUUCAAU-----UGGUGCAUUUUAAA ((((....))))((((((((..-((((((((((((((.......)))).))))))))))..)).)))))---------).(((((......-----))).))........ ( -27.90, z-score = -2.64, R) >droSim1.chrX 12673414 96 + 17042790 UGGCAAAUGUUAACCGAUGGCCCGAUUAGCUGUCAAACUGCAUAUUUGCAUAGCUAAUCAUUCAAUCGG---------UCGCCCAUUCAAU-----UGGUGCAUUUUAAA ((((....))))((((((.....((((((((((((((.......)))).)))))))))).....)))))---------).(((((......-----))).))........ ( -28.10, z-score = -2.59, R) >droSec1.super_17 301530 96 + 1527944 UGGCAAAUGUUAACCGAUGGCCCGAUUAGCUGUCAAACUGCAUAUUUGCAUAGCUAAUCACUCAAUCGG---------UCGCCCAUUCAAU-----UGGUGCAUUUUAAA ((((....))))((((((.....((((((((((((((.......)))).)))))))))).....)))))---------).(((((......-----))).))........ ( -28.10, z-score = -2.68, R) >droYak2.chrX 10584790 95 + 21770863 UGGUAAAUAUUAACCGAUUGCC-GAUUAGCUGUCCAACUGCAUAUUUGCAUAGCUAAUCACUCAAACGG---------UAGCUCAUUCAAU-----UGGUGCAUUUUAAA ............((((.(((..-((((((((((.(((........))).))))))))))...))).)))---------).(((((......-----))).))........ ( -23.10, z-score = -2.12, R) >droEre2.scaffold_4690 6836299 100 + 18748788 UGGCAAAUGGUAACCGACUGCC-GAUUAGCUGUCAAACUGCAUAUUUGCAUAGCUAAUCUCUCAAUCGGAAU----GGUCGUCCAUUCAAU-----UGGUGCAUUUUAAA .((((..(((...)))..))))-((((((((((((((.......)))).))))))))))..(((((..((((----((....)))))).))-----)))........... ( -29.90, z-score = -3.02, R) >droAna3.scaffold_13117 5345030 107 + 5790199 UGGUUA--GUUAGACAGUUACU-GGCCAAGUGUCACAGUGCAUAUUUGCAUAGGUAAUCGCUCAAUUGACAGCCCGGAUAAUGGAUCGGAUACUGGCGGUGCAUUUUUAA .(((((--((.........)))-))))(((((.(((.(((((....))))).......(((((....))(((.((((........))))...)))))))))))))).... ( -25.80, z-score = 0.35, R) >consensus UGGCAAAUGUUAACCGAUGGCC_GAUUAGCUGUCAAACUGCAUAUUUGCAUAGCUAAUCACUCAAUCGG_________UCGCCCAUUCAAU_____UGGUGCAUUUUAAA .(((.........(((((.....(((((((((......((((....))))))))))))).....)))))...........)))........................... (-15.62 = -15.73 + 0.12)

| Location | 16,478,391 – 16,478,486 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Shannon entropy | 0.35835 |
| G+C content | 0.40852 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -14.88 |
| Energy contribution | -16.11 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
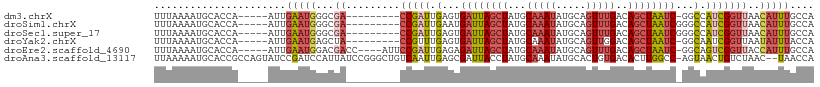
>dm3.chrX 16478391 95 - 22422827 UUUAAAAUGCACCA-----AUUGAAUGGGCGA---------CCGAUUGAGUGAUUAGCUAUGCAAAUAUGCAGUUUGACAGCUAAUC-GGCCAUCGGUUAACAUUUGCCA ........((.(((-----......)))))((---------(((((.(..(((((((((...(((((.....)))))..))))))))-).).)))))))........... ( -28.40, z-score = -2.44, R) >droSim1.chrX 12673414 96 - 17042790 UUUAAAAUGCACCA-----AUUGAAUGGGCGA---------CCGAUUGAAUGAUUAGCUAUGCAAAUAUGCAGUUUGACAGCUAAUCGGGCCAUCGGUUAACAUUUGCCA ........((.(((-----......)))))((---------(((((.(..(((((((((...(((((.....)))))..)))))))))..).)))))))........... ( -30.50, z-score = -3.07, R) >droSec1.super_17 301530 96 - 1527944 UUUAAAAUGCACCA-----AUUGAAUGGGCGA---------CCGAUUGAGUGAUUAGCUAUGCAAAUAUGCAGUUUGACAGCUAAUCGGGCCAUCGGUUAACAUUUGCCA ........((.(((-----......)))))((---------(((((.(..(((((((((...(((((.....)))))..)))))))))..).)))))))........... ( -30.00, z-score = -2.65, R) >droYak2.chrX 10584790 95 - 21770863 UUUAAAAUGCACCA-----AUUGAAUGAGCUA---------CCGUUUGAGUGAUUAGCUAUGCAAAUAUGCAGUUGGACAGCUAAUC-GGCAAUCGGUUAAUAUUUACCA ..............-----..((((((....(---------(((.(((..(((((((((.(.(((........))).).))))))))-).))).))))...))))))... ( -22.10, z-score = -1.18, R) >droEre2.scaffold_4690 6836299 100 - 18748788 UUUAAAAUGCACCA-----AUUGAAUGGACGACC----AUUCCGAUUGAGAGAUUAGCUAUGCAAAUAUGCAGUUUGACAGCUAAUC-GGCAGUCGGUUACCAUUUGCCA ....(((((.(((.-----...((((((....))----)))).(((((...((((((((...(((((.....)))))..))))))))-..))))))))...))))).... ( -31.10, z-score = -3.84, R) >droAna3.scaffold_13117 5345030 107 - 5790199 UUAAAAAUGCACCGCCAGUAUCCGAUCCAUUAUCCGGGCUGUCAAUUGAGCGAUUACCUAUGCAAAUAUGCACUGUGACACUUGGCC-AGUAACUGUCUAAC--UAACCA .......((((((((((((...((..((.......))..))...)))).)))........((((....))))..))).)).((((.(-((...))).)))).--...... ( -16.50, z-score = 1.32, R) >consensus UUUAAAAUGCACCA_____AUUGAAUGGGCGA_________CCGAUUGAGUGAUUAGCUAUGCAAAUAUGCAGUUUGACAGCUAAUC_GGCAAUCGGUUAACAUUUGCCA ...........................(((((.........(((((.(...((((((((...(((((.....)))))..))))))))...).))))).......))))). (-14.88 = -16.11 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:55 2011