
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,521,967 – 11,522,100 |
| Length | 133 |
| Max. P | 0.803315 |

| Location | 11,521,967 – 11,522,060 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.59418 |
| G+C content | 0.52725 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
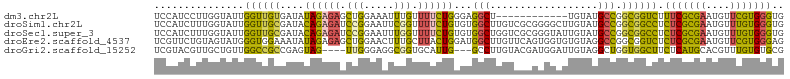
>dm3.chr2L 11521967 93 + 23011544 UCCAUCCUUGGUAUUGGUUGUGAUAUAGAGAGCUGGAAAUUUGUUUUCUGGGAGGCU------------UGUAUGCCGGCGGUCUUUCGCGAAUGUUCGUGGGUG ..(((((.(((.((...((((((...(((..(((((..((..(((((....))))).------------.))...)))))..))).))))))..))))).))))) ( -28.30, z-score = -1.54, R) >droSim1.chr2L 11339139 105 + 22036055 UCCAUCUUUGGUAUUGGUUGCGAUACAGAGAUCCGGAAUUCGGUUUUCUGUGUGGCUUGUCGCGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUG .(((....)))........((.(((((((((.(((.....))).))))))))).))..(((((..(((......))).)))))..(((((((....))))))).. ( -37.90, z-score = -1.63, R) >droSec1.super_3 6920154 105 + 7220098 UCCAUCUUUGGUAUUGGUUGCGAUACAGAGAUCCGGAAUUUGGUUUUCUGUGUGGCUGGUCGCGGGUAUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUG .(((....)))........((.(((((((((.(((.....))).))))))))).)).((((((.(((((...))))).)))))).(((((((....))))))).. ( -41.40, z-score = -3.29, R) >droEre2.scaffold_4537 34090 105 - 52796 UCGUUCUGUAGUAUGGGUGGAAAUAUAGAGAGCUGGAACUUUGCUUACUGGAUGGCUUGUUCAGUGGUGUGUAGGCCGGCGGUCUCUCGCGAAUGUUCGUGGGAG ...(((((((.............))))))).(((((..((.((((((((((((.....))))))))).))).)).)))))...(((((((((....))))))))) ( -33.72, z-score = -1.48, R) >droGri2.scaffold_15252 9353766 98 + 17193109 UCGUACGUUGCUGUUGGCCGCCGAGUAG----UUGGGAGGCGGUGCAUUG---GCCUUGUACGAUGGAUUGUAGGCUGGUGGCUUCUCAUGCACGUUUGUGUGCG .........((..(..(((..(((...(----(((.(((((.(.....).---)))))...))))...)))..)))..)..)).......(((((....))))). ( -33.20, z-score = 0.18, R) >consensus UCCAUCUUUGGUAUUGGUUGCGAUAUAGAGAGCUGGAAUUUGGUUUUCUGGGUGGCUUGUCCCGGGGAUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUG ...............((((((....((((((.(((.....))).))))))...(((..................))).)))))).(((((((....))))))).. (-14.77 = -14.25 + -0.52)

| Location | 11,522,003 – 11,522,100 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.29 |
| Shannon entropy | 0.72041 |
| G+C content | 0.59330 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -14.27 |
| Energy contribution | -13.89 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11522003 97 + 23011544 AAAUUUGUUUUCUGGGAGGCUUGU------------AUGCCGGCGGUCUUUCGCGAAUGUUCGUGGGUGCUCCACCCACGGUAUCAUGAGCUGAGUAAGCCGGCGGAGG ..((..(((((....)))))..))------------.(((((((...(((..((..(((.(((((((((...)))))))))...)))..)).)))...))))))).... ( -39.10, z-score = -2.31, R) >droSim1.chr2L 11339175 109 + 22036055 AAUUCGGUUUUCUGUGUGGCUUGUCGCGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGGAGG ..(((......((.(((((....))))).))......((((((((..(((..(((.(((.(((((((((...)))))))))...))).))).)))..))))))))))). ( -47.50, z-score = -1.72, R) >droSec1.super_3 6920190 109 + 7220098 AAUUUGGUUUUCUGUGUGGCUGGUCGCGGGUAUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGGAGG .........((((((.((((.((((((.(((((...))))).)))))).((((((.(((.(((((((((...)))))))))...))).))).)))...)))))))))). ( -49.70, z-score = -2.75, R) >droYak2.chr2L 7953793 98 + 22324452 -----------CUGGAUGCCUUUUUUUGGGGCUCGUAGGCCGGCGGAGUUUCGCGAAUGUUCGUGCGAGCUCCACUUACAGUAUCAUGAGCUGAGUACGCCGGCGGAGG -----------(((((.(((((.....))))))).)))((((((((((((((((((....))))).))))))).....((((.......)))).....))))))..... ( -41.00, z-score = -2.27, R) >droEre2.scaffold_4537 34126 109 - 52796 AACUUUGCUUACUGGAUGGCUUGUUCAGUGGUGUGUAGGCCGGCGGUCUCUCGCGAAUGUUCGUGGGAGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGGAGG ..(((((((((((((((.....))))))))).))....(((((((..(((..((..(((.(((((((.......)))))))...)))..)).)))..))))))))))). ( -47.00, z-score = -2.36, R) >dp4.chr4_group3 2404641 102 + 11692001 -------GGCUGCAUUCGGCUUGUAGGCUGGAUUGUAUGCCGGCGGACGCUCGUGGAUGUUGGUGUGGGCUCCUCCGGCCGGAUUACUGGCGGAGUAUGCGGGCGGCGG -------.(((((((((((((....))))))))..((((((((((..((....))..)))))))))).((..(((((.((((....)))))))))...))..))))).. ( -44.70, z-score = -1.03, R) >droPer1.super_1 3882306 102 + 10282868 -------GGCUGCAUUCGGCUUGUAGGCUGGAUUGUAUGCCGGCGGACGCUCGUGGAUGUUGGUGUGGGCUCCUCCGGCCGGAUUACUGGCGGAGUAUGCGGGCGGCGG -------.(((((((((((((....))))))))..((((((((((..((....))..)))))))))).((..(((((.((((....)))))))))...))..))))).. ( -44.70, z-score = -1.03, R) >droGri2.scaffold_15252 9353802 102 + 17193109 -------CGGUGCAUUGGCCUUGUACGAUGGAUUGUAGGCUGGUGGCUUCUCAUGCACGUUUGUGUGCGCUCCACCAAUCGGAUUAGAUGCCGAGUAGGCUGGCGGUGG -------.(((.(((..((((...((((....))))))))..))))))......(((((....)))))...(((((.(((......)))(((.((....)))))))))) ( -33.60, z-score = 0.47, R) >droWil1.scaffold_180772 4926678 96 - 8906247 ------------CGGCGGU-UUAUAUGAUGGAUUGUAUGCUGGAGGGCGUUCAUGUAUAUUGGUAUGAGCUCCACCCACUGGAUUUGAGACCGAAUAGGCCGGCGGUGG ------------((.((((-((((................(((..((.(((((((........))))))).))..))).(((........))).)))))))).)).... ( -28.30, z-score = -0.14, R) >consensus _______UUUUCUGUGUGGCUUGUAGGGUGGAUUGUAUGCCGGCGGACUCUCGCGAAUGUUCGUGGGAGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGGAGG ......................................((((((((...(.(((((....))))).)....)).....(((.........))).....))))))..... (-14.27 = -13.89 + -0.38)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:02 2011