
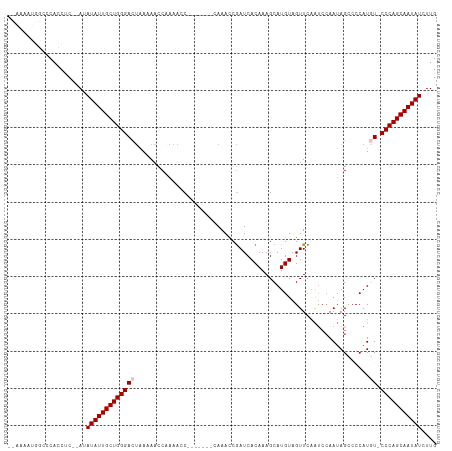
| Sequence ID | dm3.chrX |
|---|---|
| Location | 16,410,715 – 16,410,828 |
| Length | 113 |
| Max. P | 0.993510 |

| Location | 16,410,715 – 16,410,828 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Shannon entropy | 0.30518 |
| G+C content | 0.41192 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 16410715 113 + 22422827 CAAGAUAUUGCUGGG-AUAUGGGGCUAUUGGAUUGAACUACAUGCUUUGUGAUCGGUUUUUGGUUUUGGUUUUGGUUUUUAGUCCCAGCAAUAUAUAUGAGGUGGGCCAUUUUA- ....(((((((((((-((..(((((((..((((..(((((...(((........)))...))).))..)))))))))))..)))))))))))))......((....))......- ( -32.20, z-score = -2.15, R) >droSim1.chrX 12608285 105 + 17042790 CAAGAUAUUGCUGGG-ACAUGGGGCUAUUGGAUGGAACUACAUGCUUUGUGAUCGGUUUG-------GGUUUUGGUUUUUAGUCCCAGCAAUAUAU--GAGGUGGGCCAUUUUGA (((((((((((((((-((..((((((((..(((.((..((((.....)))).)).)))..-------)....)))))))..)))))))))))))..--..((....))..)))). ( -34.70, z-score = -2.88, R) >droSec1.super_17 228986 105 + 1527944 CAAGAUAUUGCUGGG-ACAUGGGGCUAUUGGAUUGAACUACAUGCUUUGUGAUUGGUUUG-------GGUUUUGGUUUUUAGUCCCAGCAAUAUAU--GAGGUGGGCCAUUUUGA (((((((((((((((-((..(((((((.....(..((((((((.....)))..)))))..-------)....)))))))..)))))))))))))..--..((....))..)))). ( -35.10, z-score = -3.32, R) >droYak2.chrX_random 376134 107 + 1802292 CAAGAUAUUGCUGGGGAAAUGGGUCUAUUGUAUUGAACUACAUGCUUUGUGAUCGAUUUG------GGUUUUGGGUUUUUAAUCCCAGCAAUAUUUGGGGGUUGGGUUGUUUG-- ..((((((((((((((....((..(((....(((((..((((.....)))).)))))...------.....)))..))....)))))))))))))).................-- ( -25.90, z-score = -1.11, R) >droEre2.scaffold_4690 8055274 107 - 18748788 CAAGAUAUUGCUGGG-ACAUGGGACUUUUUGGCUGAACUACAUGCUUUGUGAUCGGUUUA-GGUUUUGGUUUUGGUUUUAGGUCCCAGCAAUAUUU--GAGGUGGGUUUUA---- ..(((((((((((((-((.(((((((..(..(((((((((((.....)))....))))))-)...)..)....))))))).)))))))))))))))--.............---- ( -35.20, z-score = -3.89, R) >consensus CAAGAUAUUGCUGGG_ACAUGGGGCUAUUGGAUUGAACUACAUGCUUUGUGAUCGGUUUG_______GGUUUUGGUUUUUAGUCCCAGCAAUAUAU__GAGGUGGGCCAUUUU__ ....(((((((((((.((..(((((((.((((((((..((((.....)))).))))))))............)))))))..)))))))))))))..................... (-21.92 = -22.52 + 0.60)


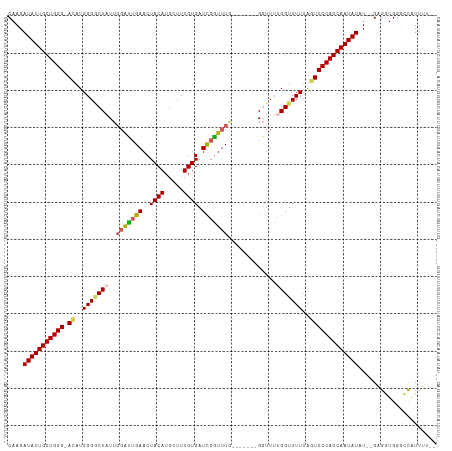
| Location | 16,410,715 – 16,410,828 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Shannon entropy | 0.30518 |
| G+C content | 0.41192 |
| Mean single sequence MFE | -18.72 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
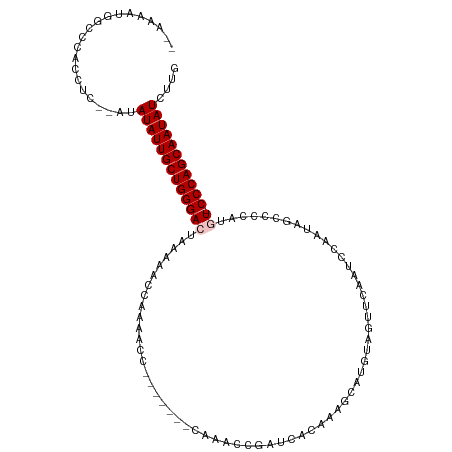
>dm3.chrX 16410715 113 - 22422827 -UAAAAUGGCCCACCUCAUAUAUAUUGCUGGGACUAAAAACCAAAACCAAAACCAAAAACCGAUCACAAAGCAUGUAGUUCAAUCCAAUAGCCCCAUAU-CCCAGCAAUAUCUUG -....................((((((((((((............................((.((((.....))).).)).......((......)))-))))))))))).... ( -16.70, z-score = -1.77, R) >droSim1.chrX 12608285 105 - 17042790 UCAAAAUGGCCCACCUC--AUAUAUUGCUGGGACUAAAAACCAAAACC-------CAAACCGAUCACAAAGCAUGUAGUUCCAUCCAAUAGCCCCAUGU-CCCAGCAAUAUCUUG .................--..(((((((((((((..............-------......(....).....(((..(((.........)))..)))))-))))))))))).... ( -19.60, z-score = -2.39, R) >droSec1.super_17 228986 105 - 1527944 UCAAAAUGGCCCACCUC--AUAUAUUGCUGGGACUAAAAACCAAAACC-------CAAACCAAUCACAAAGCAUGUAGUUCAAUCCAAUAGCCCCAUGU-CCCAGCAAUAUCUUG .................--..(((((((((((((..............-------..........(((.....))).(((.........))).....))-))))))))))).... ( -18.90, z-score = -2.34, R) >droYak2.chrX_random 376134 107 - 1802292 --CAAACAACCCAACCCCCAAAUAUUGCUGGGAUUAAAAACCCAAAACC------CAAAUCGAUCACAAAGCAUGUAGUUCAAUACAAUAGACCCAUUUCCCCAGCAAUAUCUUG --................((((((((((((((.................------..................((((......))))...((......))))))))))))).))) ( -18.00, z-score = -3.94, R) >droEre2.scaffold_4690 8055274 107 + 18748788 ----UAAAACCCACCUC--AAAUAUUGCUGGGACCUAAAACCAAAACCAAAACC-UAAACCGAUCACAAAGCAUGUAGUUCAGCCAAAAAGUCCCAUGU-CCCAGCAAUAUCUUG ----............(--(((((((((((((((....................-......((.((((.....))).).)).((......)).....))-))))))))))).))) ( -20.40, z-score = -4.13, R) >consensus __AAAAUGGCCCACCUC__AUAUAUUGCUGGGACUAAAAACCAAAACC_______CAAACCGAUCACAAAGCAUGUAGUUCAAUCCAAUAGCCCCAUGU_CCCAGCAAUAUCUUG .....................(((((((((((.............................(((.(((.....))).))).......(((......))).))))))))))).... (-13.72 = -13.88 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:49:47 2011